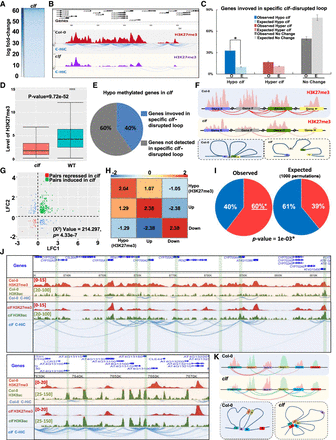
Reduction of H3K27me3 levels induces a reconfiguration of chromatin architecture. (A) Heatmap of C-Hi-C data showing loops that are weaker in clf mutant than in wild type. (B) Example of C-Hi-C interactions displaying disrupted loops in clf compared to wild type. C-Hi-C interactions (blue lines) and H3K27me3 ChIP-seq signal in wild type (red peaks) and clf (purple peaks) are represented. (C) Histogram representing the percentage of genes (observed [O] or expected [E]) involved in cSDLs that are either hyper- or hypomethylated in clf compared to WT. To obtain the expected proportion, we shuffled the H3K27me3 signals 1000 times to obtain the randomized gene counts. The mean of the 1000 permutations was used to determine the expected proportions. Asterisk indicates significant difference (P-value < 2.2 × 10−16, test of proportions). (D) The box plot displays the H3K27me3 levels of the 40% of clf hypomethylated genes involved in cSDLs. (E) The pie chart represents the percentage of clf hypomethylated genes involved in cSDLs. (F) Model of chromatin contacts organization in wild type and clf mutant. (G) Scatterplot of log2 (clf/wild-type gene expression fold change) for pairs of genes interacting specifically in wild type compared to clf. (H) Heatmap presenting the log2 of odd ratios of combinations of features of interacting genes (Results). Positive log2 (odd ratio) indicates enrichment and negative indicates depletion. (I) Pie chart representing the proportion of loops involving a gene H3K27me3 hypomethylated and a gene marked or not by H3K9ac in clf mutant. (Hypo-H3K27me3) No H3K9ac loops in clf mutant (blue: 40% observed and 61% expected, respectively); (Hypo-H3K27me3) H3K9ac loops (red: 60% observed and 39% expected, respectively). (J) Examples of C-Hi-C interactions of a region losing H3K27me3 in clf and that tend to establish interactions with regions marked with H3K9ac euchromatin histone modification. C-Hi-C interactions (blue lines), H3K9ac ChIP-seq signal in wild type and clf (green peaks), H3K27me3 ChIP-seq signal in wild type and clf (red peaks) are represented. (K) Model of chromatin contacts organization in wild type and clf mutant.











