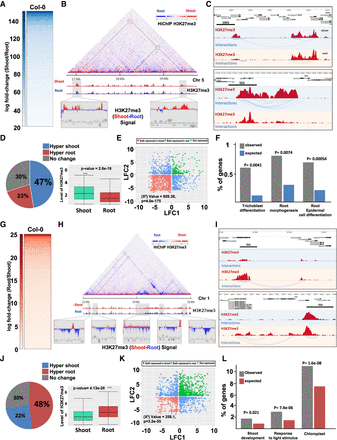
Shoot and root nuclei display distinct 3D chromatin architectures. (A) Heatmap showing the H3K27me3 HiChIP signal of the top shoot-specific repressive loops (SSRLs). (B) Example of long-distance SSRL on Chromosome 5. ChIP-seq signals of H3K27me3 in shoot (red peaks) and root (blue peaks) were aligned with the map and the differential analysis of both ChIP-seq signals in differentially interacting regions are highlighted (bottom panels in gray). (C) Example of short distance SSRL. H3K27me3 ChIP-seq signal is represented by red peaks and chromatin interactions signal by blue lines. (D) Analysis of H3K27me3 levels on SSRLs. The pie chart represents the percentage of the genes involved in SSRLs that are either hypermethylated in shoot or in root. The box plot shows the H3K27me3 levels in shoot or root of the 47% of shoot hypermethylated genes involved in SSRLs. (E) Scatterplot of log2 (shoot/root gene expression fold change) for pairs of genes interacting through H3K27me3-associated contacts in shoot. (F) Gene Ontology enrichment analysis of the differentially expressed genes involved in SSRLs. (G) Heatmap of H3K27me3 HiChIP signal of the top root-specific repressive loops (RSRLs). (H) Example of long-distance RSRLs on Chromosome 1. ChIP-seq signals of H3K27me3 in shoot (red peaks) and root (blue peaks) were aligned with the map and the differential analysis of both ChIP-seq signals in differentially interacting regions are highlighted (bottom panels in gray). (I) Example of short distance RSRLs. (J) Analysis of H3K27me3 level over RSRLs. The pie chart represents the percentage of genes involved in RSRLs that are either hypermethylated in shoot or root. The box plot displayed the H3K27me3 levels of the 48% of root hypermethylated genes involved in RSRLs. (K) Scatterplot of log2 (shoot/root gene expression fold change) for pairs of genes interacting through H3K27me3-associated contacts in root. (L) Gene Ontology enrichment analysis of the differentially expressed genes involved in RSRLs.











