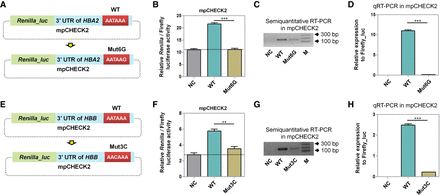
Investigation on the effect of PAS variants occurring in patients using the mpCHECK2 system. (A) The sixth position of the PAS in HBA2 was changed from A (WT) to G (Mut6G) in mpCHECK2. (B) Comparison of normalized Renilla luciferase activity in HUVEC transfected with mpCHECK2 constructs of NC, WT, and Mut6G (panel A). (C,D) Comparison of mRNA expression levels in HUVEC transfected with vectors in panel A (NC, WT, and Mut6G) using both semiquantitative PCR (C) and qRT-PCR (D). (E) Mutations in the third PAS position of HBB (T → C) in mpCHECK2. (F) Comparison of normalized Renilla luciferase activity in HUVEC transfected with mpCHECK2 constructs of NC, WT, and Mut3C (panel E). (G,H) Comparison of mRNA expression levels in HUVEC transfected with NC, WT, and Mut3C (panel E) using semiquantitative PCR (G) and qRT-PCR (H). All luciferase activity assays were performed with four replicates and all qRT-PCR reactions were carried out with three replicates. Data were presented as mean ± SEM. (***) P < 0.001, (**) P < 0.01; one-way ANOVA test.











