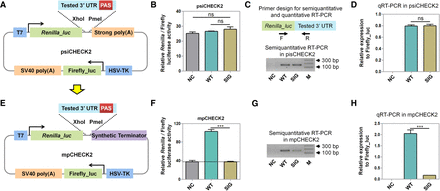
The establishment of an mpCHECK2 system to evaluate the effect of PAS variants. (A) The schematic diagram of the original psiCHECK2 vector with a highly efficient synthetic PAS downstream of the tested 3′ UTR (labeled strong polyA). (B) Renilla luciferase activity (normalized by firefly) in HUVEC transfected using psiCHECK2 without a tested 3′ UTR (NC), with the 3′ UTR of HBA2 containing a WT PAS (AATAAA) (WT), and a 3′ UTR of HBA2 containing a mutated PAS (GGATCC) (SIG), respectively. (C,D) Comparison of mRNA expression levels among NC, WT, and SIG (based on psiCHECK2, described in panel B, that was transfected into HUVEC cells using both semiquantitative PCR (C) and qRT-PCR (D). Primer design is illustrated on the top and M denotes molecular size markers. (E) The modified mpCHECK2 vector resulted from replacing the strong PAS in psiCHECK2 with a highly efficient synthetic terminator. (F) Comparison of normalized Renilla luciferase activity in HUVEC transfected with mpCHECK2 vector (panel E) of NC, WT, and SIG, as described above. Dashed line indicates the basal luciferase signal of NC. (G,H) Comparison of mRNA expression levels in HUVEC transfected with NC, WT, and SIG (based on mpCHECK2) using semiquantitative PCR (G) and qRT-PCR (H). All luciferase activity assays were performed with four replicates and all qRT-PCR reactions were carried out with three replicates. Data are presented as mean ± SEM. (***) P < 0.001, (ns) not significant; one-way ANOVA test.











