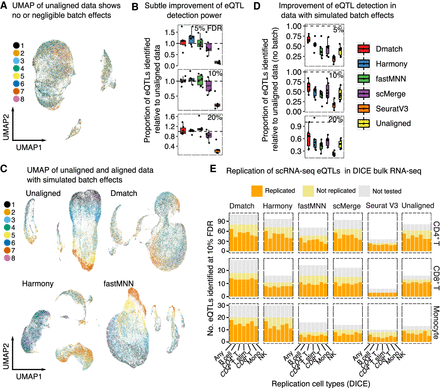
Evaluation of alignment methods on eQTL mapping. (A) UMAP of unaligned PBMC scRNA-seq data from 45 donors (van der Wijst et al. 2018). The cells from the 45 donors were pooled into eight samples for sequencing. (B) Alignment slightly improves mapping power. (C) UMAP of unaligned PBMC data with spiked-in batch effects (see Methods). (D) Alignment improves eQTL mapping power substantially when batch effects are simulated to reflect a realistic and practical scRNA-seq collection strategy. (E) Replication of eQTLs identified in a bulk RNA-seq study of immune cell type eQTLs from the DICE consortium. The eQTLs that were not tested correspond to SNPs whose genotype could not be determined or to genes filtered out due to low expression.











