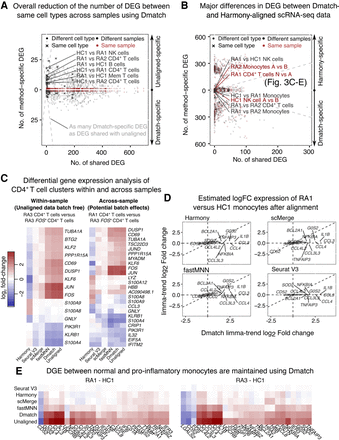
Evidence for overcorrection in aligned scRNA-seq data. (A) Scatterplot showing the number of differentially expressed genes (DEGs) that are identified using unaligned data (positive y-axis) or using data aligned using Dmatch (negative y-axis), versus the number of DEGs that are identified in both data sets (x-axis). Each point represents a comparison between cell type clusters from the same or different cell type, or from the same or different sample. Successful removal of batch effects is supported by smaller numbers of DEGs resulting from Dmatch-aligned data in same cell type comparisons, compared to unaligned data. DEGs inferred from within-sample cluster comparisons are identical between unaligned and Dmatch-aligned data. (B) Scatterplot similar to A shows a large difference between Dmatch and Harmony-aligned data. (C) Heat maps of estimated DEG fold changes between CD4+ T cells versus CD4+ T cells expressing FOS within the same sample (RA3, left heat map), and across two samples (RA1 vs. RA3, right heat map). Dmatch and fastMNN estimates of DEG fold changes within samples are consistent with unaligned data, as we should expect. However, the estimates from scMerge and Harmony are shrunken and inconsistent, respectively, suggesting overcorrection. Across-sample comparison between the two cell types shows an increase in JUN, FOS, and CD69 expression in activated CD4+ T cells, consistent with within-sample comparisons for Dmatch and fastMNN. The signal is reduced or reversed for scMerge and Harmony, respectively. (D) Scatterplot of DEG log fold change comparison between CD14+ monocytes from healthy individual HC and RA patient (RA1). Fold change estimates are reduced or zero for inflammatory markers (IL1B, CCL3, CCL4) using data obtained from fastMNN, Harmony, and scMerge. (E) Heat maps of estimated DEG fold change comparison between RA1-HC1 and RA3-HC1 are consistent and support the presence of overcorrection in data aligned using Harmony, scMerge, and fastMNN, masking real biological signal.











