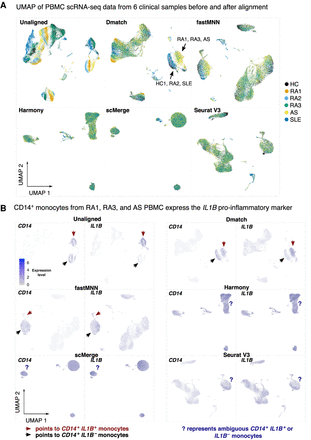
Application of alignment tools on PBMC clinical samples. (A) UMAP dimensionality reduction of PBMC scRNA-seq data from six clinical samples before and after alignment comparing Dmatch, fastMNN, Harmony, scMerge, and Seurat V3. UMAP plots suggest possible overcorrection from scMerge. (B) We observed that CD14+ monocytes express IL1B in a subset of individuals with autoimmune disease (RA1, RA3, AS). This signal was preserved after alignment using Dmatch and, to some extent, fastMNN, whereas it disappeared using Harmony, Seurat V3, and scMerge. Shades of purple represent marker expression in each cell relative to other cells in the same UMAP.











