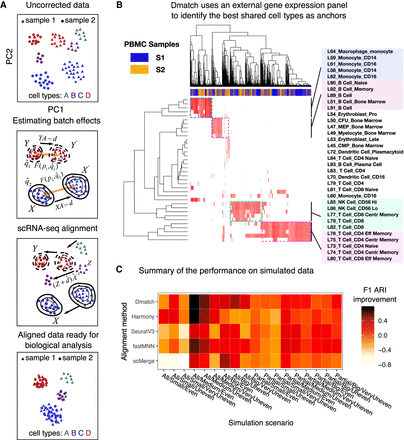
Overview of Dmatch and simulations. (A) Data processing pipeline. First, the uncorrected data are projected onto principal components (PC). Next, an external gene expression panel is used to identify anchor cells to estimate linear batch effects in the form of a rotation and a translation in PC space. Last, the data are corrected by rotating and translating the data points in PC space. The PC loadings are used to recover the aligned data to allow downstream analyses. (B) Dmatch uses a large reference transcriptomes from the Primary Cell Atlas to identify subpopulations from the observed cells based on the projection. These subpopulations are used as anchors to guide the alignment. We show an example applied on real data, which demonstrates the identification of cell clusters corresponding to monocytes, B cells, and two different subclasses of T cells. (C) Heat map showing the ARI F1 scores (Methods) improvements for stimulated data corrected using different alignment methods over unaligned simulated data. Simulations were based on real PBMC data that were split into two batches (see Methods) such that: (1) all cell types were shared or partially shared (All or Partial); (2) noise was added to simulate small, medium, and large batch effect sizes (Small, Medium, or Big); and (3) data were split into two batches such that the cells from each cell type were distributed evenly, unevenly, or very unevenly across the two batches (ratios of 1:1, 1:2, or 1:5, for Even, Uneven, and VeryUneven, respectively). The overall performance of Dmatch as measured by ARI F1 scores was the best overall, followed by Harmony, and fastMNN.











