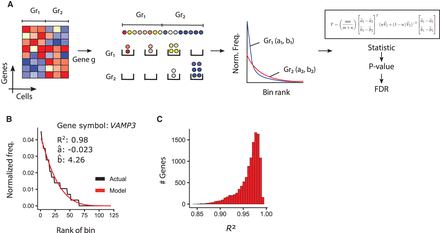
Modeling single-cell gene expression using ROSeq. (A) As part of the ROSeq differential expression analysis workflow, cells are first binned depending on expression values associated with a particular gene. For each cell-group, bins are ranked depending on cell frequency. The discrete generalized beta distribution (DGBD) is used as a probability mass function to express a normalized bin-wise cell frequency as a function of its corresponding rank using two real parameters a and b. A Wald-type test is used on the MLE of these parameters across the cell-groups to find differentially expressed genes. (B) DGBD-based modeling of VAMP3 expression (source: Tung data) (Tung et al. 2017). Discretized expression bins are ranked based on normalized bin-wise cellular frequencies. (C) Distribution of R2 values obtained from DGBD-based modeling of 11,513 expressed genes (source: Tung data) (Tung et al. 2017).











