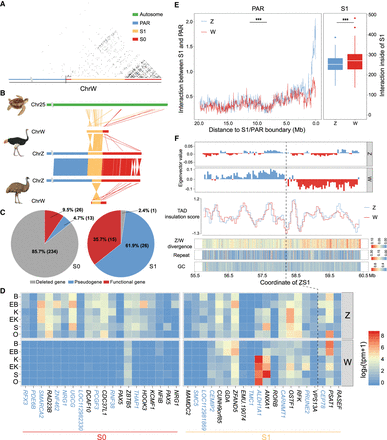
Emu sex chromosome evolution. (A) Dot plot of SDR of Chr W, which is segregated into a highly repetitive S0 (red) and only moderately repetitive S1 (orange). Part of the S0 region was reshuffled between the PAR (blue) and the S1, which was inferred based on its homology with the Z-linked S0 region (Fig. 1E). (B) Syntenic plot between the ZW Chromosomes of emu and ostrich and the homologous autosome of green sea turtle, which suggests a W-linked inversion created the emu S1. (C) There are more genes that have become deleted (gray) or pseudogenes (blue, those containing premature stop codons or frameshift mutations) in S0 than in S1 on the W Chromosome. (D) The log expression levels (TPM) of single-copy homologous genes in the S0 and S1 regions of Z and W Chromosomes. (B) Brain, (EB) embryonic brain, (K) kidney, (EK) embryonic kidney, (S) spleen, (O) ovary. The genes with blue color indicate the pseudogenes. (E) The left panel plot shows the cis-contacts between the PAR and the Z- (blue) and W- (red) linked S1 regions, which exhibit reduced W-linked contacts compared to the Z-linked contacts. The x-axis shows the distance to the S1/PAR boundary. The right panel plot indicates higher cis-contacts within the W-linked S1 region compared to its homologous Z-linked region. (***) P < 0.0005. (F) The W-linked S1 is segregated into two compartments WS1A (left, blue) and WS1B (right, red). From upper to lower: A(blue)/B(red) compartment division based on the Hi-C contact profiles; TAD insulation scores (the lower values correspond to the TAD boundaries); ZW pairwise sequence divergence level, repeat, and GC content. The black line indicates the boundary (58.22 Mb on Chr Z and 5.28 Mb on Chr W) between WS1A and WS1B. Note: The WS1 has been reversed for the convenience of ZW comparison.











