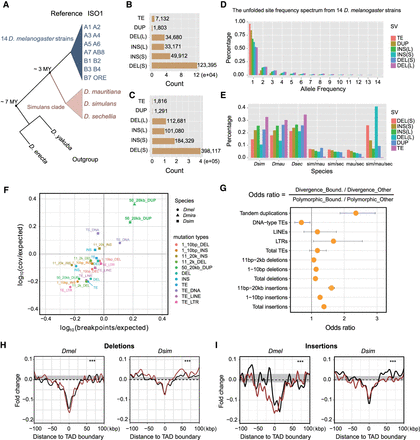
Patterns of structural variants at Drosophila TAD boundaries. (A) Highly contiguous genome assemblies from 14 D. melanogaster strains and three D. simulans clade species, together with two outgroup species, D. erecta and D. yakuba. (B) Nonredundant SVs, including TE insertions, tandem duplications (DUP), non-TE insertions (INS; “S” represents insertions size range from 1-10 bp and “L” represents insertions size range from 11 bp to 20 kbp), and deletions (DEL; “S” for 1–10 bp and “L” for 11 bp to 2 kbp) identified from the 14 D. melanogaster strains. (C) Nonredundant SVs identified in the three D. simulans clade species. (D) The unfolded site frequency spectrum of SVs from 14 D. melanogaster strains. (E) Phylogenetic profiling of SVs among the three D. simulans clade species. (F) Tests of purifying selection on SVs at TAD boundaries using Fudenberg and Pollard's method. (G) Odds ratios of 2 × 2 contingency tables with margins categorizing polymorphism/divergence and boundary/nonboundary mutations. Confidence intervals are calculated from the Fisher's exact test results. (H) Deletions from both data sets are depleted at the TAD boundaries. (I) Non-TE insertions from both data sets are depleted at the TAD boundaries. Red and black lines represent larger and shorter variants, respectively. (***) P < 1 × 10−4, permutation test (Supplemental Fig. S15).











