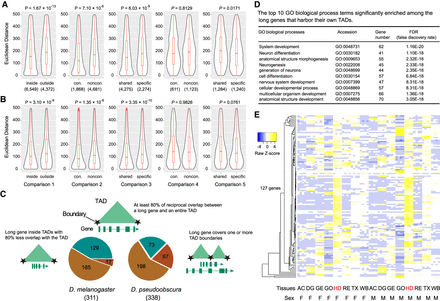
The potential roles of TADs in gene regulation in Drosophila. (A) Expression divergence measured by Euclidean distance for one-to-one orthologs between D. melanogaster and D. pseudoobscura. (B) Expression variation measured by Euclidean distance for the same gene sets used in the above interspecific comparison between two D. melanogaster strains, OreR and w1118. (C) Physical overlap of long genes and TADs in D. melanogaster and D. pseudoobscura. (D) Representative GO biological process terms significantly enriched among the 127 long genes that constitute their own TADs in D. melanogaster. (E) Expression profile of the 127 long genes across eight tissues in both male and female in D. melanogaster. (AC) Abdomen without digestive or reproductive system; (DG) digestive plus excretory system; (GE) genitalia; (GO) gonad; (HD) head; (RE) reproductive system without gonad; (TX) thorax without digestive system; (WB) whole body; (F) female; (M) male.











