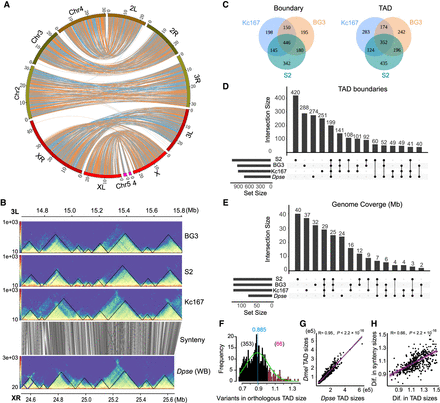
Evolutionary conservation of TADs between D. pseudoobscura (Dpse) and D. melanogaster (Dmel). (A) Genome synteny map between Dmel and Dpse constructed using 985 syntenic blocks >10 kbp. (B) Conservation of TADs on a ∼1.2-Mb orthologous region between Dmel and Dpse. (C) Overlap of TAD features across three D. melanogaster cell lines. (D) Upset plot showing the overlap of TAD boundaries across three D. melanogaster cell lines and D. pseudoobscura whole body; 5-kbp boundaries for Dmel and 10-kbp boundaries for Dpse. (E) Upset plot showing genome coverage that maintained conserved TADs across three D. melanogaster cell lines and D. pseudoobscura. (F) Distribution of size variation (Dmel size divided by Dpse size) of orthologous TADs between Dmel and Dpse. (G) Correlation of the size of conserved TADs between Dmel and Dpse. (H) Correlation between the size difference of the orthologous TADs and the size difference of local synteny blocks where the orthologous TADs are located between Dmel and Dpse. (Dif.) Difference.











