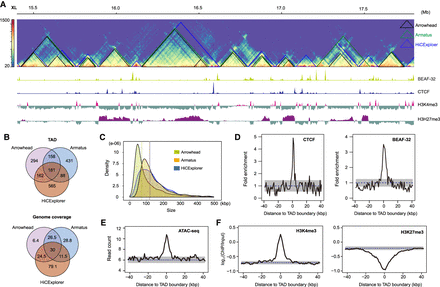
TADs annotated using full body samples in D. pseudoobscura. (A) Hi-C contact map (5-kbp resolution) on a ∼3-Mb region from the X Chromosome with TADs annotated using Arrowhead (black), Armatus (green), and HiCExplorer (blue). The bottom browser tracks show the local profiles of binding sites of BEAF-32 and CTCF, as well as two histone marks (H3K4me3 and H3H27me3). (B) Overlap of TADs and their genome coverage annotated using three tools. (C) TAD size distribution for three tools. Vertical dashed lines represent the mean values. (D) Enrichment of CTCF and BEAF-32 binding sites at HiCExplorer TAD boundaries (P < 1.0 × 10−4). (E) Enrichment of ATAC-seq signal (open chromatin marks) at HiCExplorer TAD boundaries (P < 1.0 × 10−4). (F) TAD boundaries (HiCExplorer) are enriched in H3K4me3 (P < 1.0 × 10−4) but depleted for H3K27me3 marks (P < 1.0 × 10−4). P-values (D–F) were determined by permutation tests (n = 10,000); dashed lines represent mean values obtained from permutation tests; gray shaded areas, mean ± SD in D and 95% intervals in E and F.











