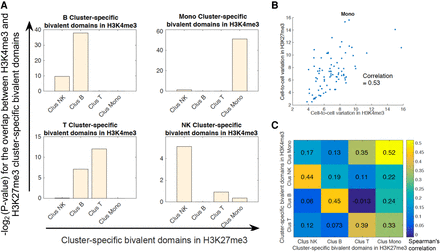
Correlation of cell clusters revealed from the single-cell H3K4me3 and H3K27me3 data by bivalent domains. (A) The cluster-specific peaks identified from the single-cell H3K4me3 and H3K27me3 data exhibit the highest overlap if they are from the same cell type. For each subplot, the cluster-specific peaks of H3K4me3 from one annotated cluster (as indicated on the top) were compared with the cluster-specific peaks of H3K27me3 from different clusters (as indicated below the plot). The y-axis in each subplot indicates the −log2 of P-value for the overlap between the cluster-specific peaks of H3K4me3 and cluster-specific peaks of H3K27me3. (B) A scatterplot between the cell-to-cell variation of H3K4me3 and H3K27me3 for clusters annotated as monocytes in bivalent domains (Methods). (C) Cluster-specific bivalent domains associated with H3K4me3 and H3K27me3 were computed for the purpose of finding the relationship between cell-to-cell variation in H3K4me3 and H3K27me3. For each comparison between the H3K4me3 and H3K27me3 clusters, the overlap between cluster-specific bivalent domains was considered; the Spearman's correlation between the coefficient of variation in H3K4me3 and H3K27me3 for these selected bivalent domains was calculated.











