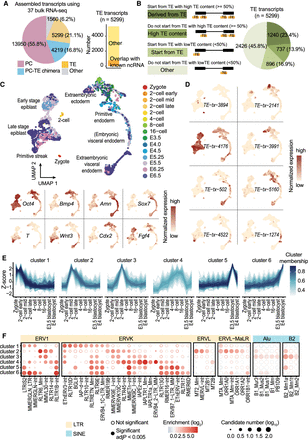
Dynamic TE expression in mouse preimplantation embryos. (A) Using 37 bulk RNA-seq samples, 5299 TE transcripts were constructed. Of these, 770 TE transcripts overlap with ncRNAs annotated by RefSeq. (B) More than half of all the assembled TE transcripts either initiate from TEs or have >50% of their exons composed of TEs. (C, upper) UMAP of scRNA-seq data from mouse zygote to E6.5 embryos. Cells were colored based on developmental stages. (Lower) Expression of cell type–specific markers. (D) Examples of developmental stage– and tissue-specific TE transcripts. (E) TE transcripts were grouped into six clusters based on their expression pattern across preimplantation stages. (F) TE subfamily enrichment analysis using TE transcripts within each of the six clusters.











