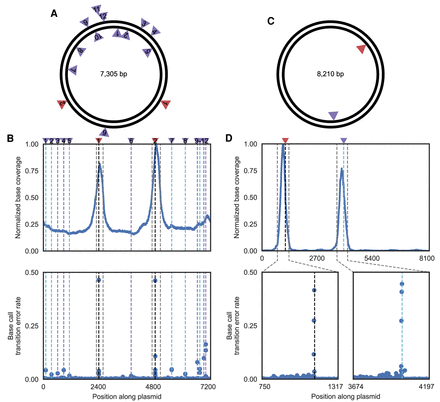
Detection of off-target nicks in plasmid DNA. (A) Schematic of a plasmid used to test DENT-seq, in which red arrows represent on-target activity of Nb.BsmI and purple arrows represent detected off-target activity of Nb.BsmI. (B) Normalized sequence coverage (top) and base transition rate (bottom) for the plasmid in A. Black dashed lines represent on-target locations of Nb.BsmI activity and gray dashed lines represent the bounds of called MACS2 peaks. Purple and cyan dashed lines represent detected off-target locations of Nb.BsmI activity on the reference and non-reference strands, respectively. (C) Schematic of a plasmid used to test DENT-seq in which the red arrow represents on-target activity of D10A spCas9 nickase and the purple arrow represents detected off-target activity (both are on the non-reference strand). (D) Normalized sequence coverage (top) and base transition rate (bottom) for the plasmid in C. The black dashed line represents on-target activity of D10A spCas9 nickase, whereas the cyan dashed line represents off-target activity. The gray dashed lines represent the bounds of called MACS2 peaks.











