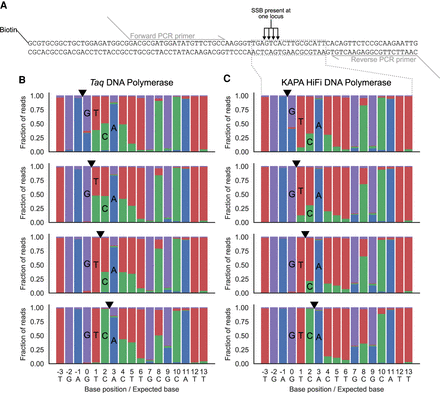
Effect of degenerate nucleotides during PCR. (A) Schematic of the oligonucleotides used to test the mutational outcome when dPTP and dKTP are incorporated into DNA and amplified by PCR. Four different oligonucleotides were generated, each with a nick directly 5′ of a different one of the four native dNTPs. The strand with the nick also contained a 5′ biotin-TEG modification to allow for purification of just that strand before PCR. PCR primers were designed to specifically amplify the region of the oligonucleotides containing the nicks and were tailed with 5′ sequences compatible with secondary PCR using barcoded P5 and P7 sequencing primers. (B,C) Sequencing result of the four oligonucleotides following consecutive nick translations with dPTP plus dKTP and then with standard dNTPs, purification, and PCR with Taq DNA polymerase (B) or KAPA HiFi DNA polymerase (C). The black triangles represent the location of the nick for each sample.











