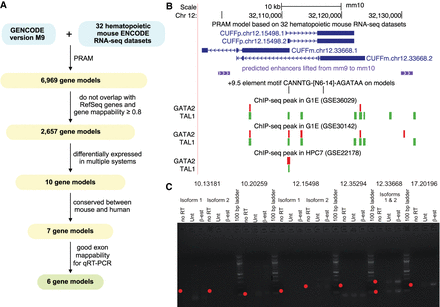
Genomic features and experimental validations of PRAM mouse transcripts. (A) Workflow of applying PRAM to discover transcripts from mouse hematopoiesis-related RNA-seq data sets: input (cyan), intermediate results (yellow), and output (green). (B) PRAM transcripts CUFFp.chr12.15498 and CUFFm.chr12.33668 had multiple supporting genomic features from external data sets. (C) Semi-qRT-PCR measurements of the six PRAM models in untreated (Unt) and 48-h ß-estradiol (ß-est)-treated G1E-ER-GATA1 cells. Red dots demarcate anticipated transcript sizes. Isoforms with splice junctions distant from each other were measured separately. Gene model name prefixes were removed for brevity.











