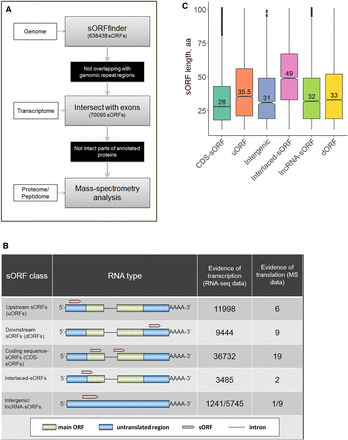
Several distinct types of sORFs are present in the moss genome. (A) Pipeline used in this study to identify coding sORFs. (B) Proposed classification of sORFs according to the types of encoding transcripts: upstream ORFs (uORFs) and downstream ORFs (dORFs) in the untranslated regions (UTRs) of canonical mRNAs; CDS-sORFs, which overlap with protein-coding sequences in alternative (+2 or +3) reading frames or are truncated versions of proteins generated by alternative splicing; interlaced-sORFs, which overlap both the protein-coding sequence and UTR on the same transcript; lncRNA-sORFs and intergenic sORFs, which are located on short nonprotein-coding transcripts. (C) Box plot of the length distribution of sORFs in different groups.











