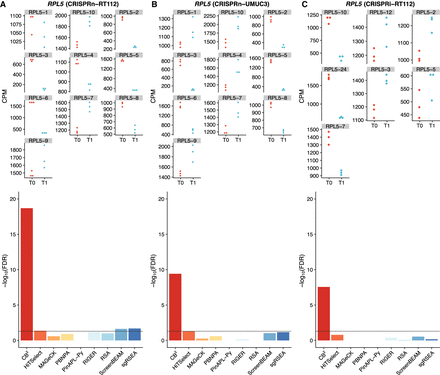
CB2 detects essential genes missed by other leading methods: the case of RPL5. sgRNA quantification for RPL5 in cell line (A) RT112, (B) UMUC3 using CRISPRn, and (C) RT112 using the CRISPRi library. The top panels show counts per million (CPM) of sgRNAs that target RPL5 for each group (T0 and T1), and the bottom panels indicate the reported the FDR for RPL5 in each screen across all the methods. A horizontal line at FDR = 0.01 is used as a threshold for statistical cutoff. CB2 outperforms all other methods of identifying RPL5 as an essential gene across all benchmark data sets.











