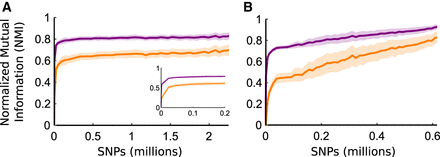
Normalized mutual information (NMI) between the PST inferred using the entire genome and PSTs inferred from subsampled fractions of the genome. The NMI values evaluate the amount of information gained on hierarchical population structure by sampling a fraction of the data set. The mean NMI across 100 random subsamples for each SNP coverage value considering the entire PST topology is shown in purple; mean NMI considering only the finest-scale leaf clusters is shown in orange. Shaded regions show standard deviations across 100 sampling replicates. (A) A. thaliana data set. Inset shows NMI for subsamples below 200,000 SNPs. (B) Human data set. NMI values saturate at values below 1 because cluster assignments often switch at fine scales (e.g., between PST leaves) when PSTs are inferred from subsampled data.











