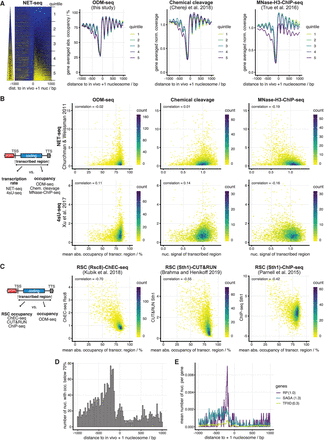
Correlation of absolute occupancy with biological features. (A, left) In vivo +1 nucleosome-aligned heat map of NET-seq data monitoring nascent RNA bound to RNA polymerase (Churchman and Weissman 2011) sorted from top to bottom by increasing signal over the gene body. (Right) As in Figure 2B but for the indicated data sets and genes subdivided according to quintiles of sorting in heat map on the left. (B) Correlation plots (color indicates number of occurrences) of transcription rate (NET-seq as in A or 4sU-seq [Xu et al. 2017]) against the absolute occupancy or coverage averaged over transcribed regions for the indicated data sets as in A. (C) As in B but correlation of absolute occupancy averaged over transcribed regions with RSC binding measured by the indicated methods. (D) +1 Nucleosome-aligned histogram (accumulated in 20-bp bins) of nucleosomes dyads (Chereji et al. 2018) with <70% absolute occupancy. (E) As in D but clustered by gene groups (Vinayachandran et al. 2018) as indicated. In brackets, mean number of low absolute occupancy nucleosomes per gene in 2-kb window around in vivo +1 nucleosome. Used data sets are listed in Supplemental Table S2.











