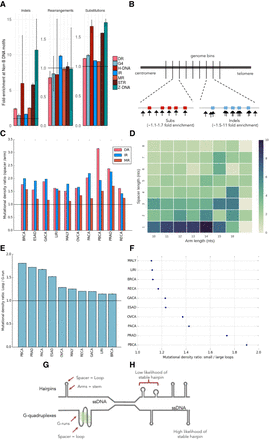
Non-B DNA motifs are mechanistically linked to mutability through formation of secondary structures. (A) Enrichment of mutagenesis for non-B motifs within their genomic bins, thus correcting for genomic GC variation. Error bars represent the standard error. (B) Depiction of enrichment per genomic bin, for results in A, demonstrating how mutations are enriched for non-B motifs. Red and blue boxes represent non-B motifs. (C) Mutational density in spacers compared to arms for direct repeats, inverted repeats, and mirror repeats across 10 tumor types. Error bars representing standard error are too small to visualize. A Wilcoxon signed-rank test was performed (P-value < 0.001 across all tumors for IR, MR, DR). (D) Heat map showing relative ratio of mutational density of spacers over arms for breast cancer at inverted repeats. (E) Enrichment of mutation density in loops: G-runs across ten cancer types. Error bars represent standard error from bootstrapping with replacement (n = 10,000). (F) Enrichment of mutation density at G-quadruplexes for small loop sizes (≤3 nt) relative to large loop sizes (>3 nt) across 10 cancer types. Error bars represent standard error from bootstrapping with replacement (n = 10,000). A Mann-Whitney U test was performed for each cancer type (P-value < 0.001 across all tumor types). (G) Depiction of two very different secondary structures that both have loop domains which are more mutable than their other components. (H) Some non-B motifs have characteristics such as arm or spacer lengths that increase the likelihood of stable hairpin formation. These perhaps can occur stably more frequently, and thus, their exposed regions are more likely to be damaged and mutated.











