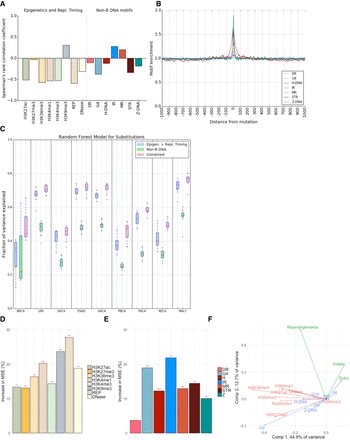
Non-B DNA motifs predict somatic mutability in human cancers. (A) Correlations between the number of non-B DNA motifs, and epigenetic features and replication timing, with the number of substitutions (Spearman's rank correlation coefficient). Please note interpretation is directional, e.g., a positive correlation with replication time would indicate increased mutability with early replication time domains, while a negative correlation denotes increased mutability in late replication time domains. (B) The distribution of different non-B DNA motifs in a window of 2 kb centered on substitutions across all tumor types. (C) Fraction of variance explained for predicting the number of mutations in 500-kb bins with random forest regression using non B-DNA motifs and epigenetic features/replication timing as predictors for multiple tumor types. (BRCA) Breast cancer, (LIRI) liver cancer, (OVCA) ovarian cancer, (ESAD) esophageal adenocarcinoma, (GACA) gastric cancer, (PBCA) pedriatic brain cancer, (PACA) pancreatic cancer, (RECA) renal cell carcinoma, (MALY) malignant lymphoma. Error bars represent standard error from 10-fold cross-validation. (D,E) Importance of the different predictors for the random forest regression. The y-axis shows the increase in mean square error (MSE) when the variable is excluded. (**) FDR < 0.01, as determined by a permutation test. (F) PCA. The first two principal components separate mutations (green), non-B DNA motifs (blue), and epigenetics and replication timing domains (red).











