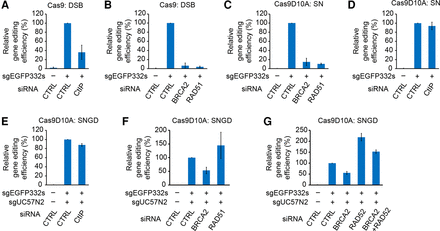
Gene-editing efficiency in HDR factor-depleted cells. The relative gene correction efficiency in the indicated siRNA-transfected cells (relative to the gene-editing efficiency in siCTRL-transfected cells; mean ± SD, N = 3) is presented in each panel. m332pamPD was used as a donor for all experiments. (A,B) DSB-mediated gene editing in CtIP-, BRCA2-, or RAD51-depleted HeLa EGFPcC>G reporter cells. A DSB was introduced at the sgEGFP332s target site of the reporter. (C,D) SN-mediated gene editing in BRCA2-, RAD51-, or CtIP-depleted HeLa EGFPcC>G reporter cells. A nick was introduced at the sgEGFP332s target site of the reporter. (E,F) SNGD-mediated gene editing in CtIP-, BRCA2-, or RAD51-depleted EGFPcC>G reporter cells. (G) SNGD-mediated gene editing in RAD52- and/or BRCA2-depleted HeLa EGFPcC>G reporter cells. Nicks were introduced at the sgEGFP332s target site of the reporter and at the sgUC57N2 site of the donor plasmid for SNGD.











