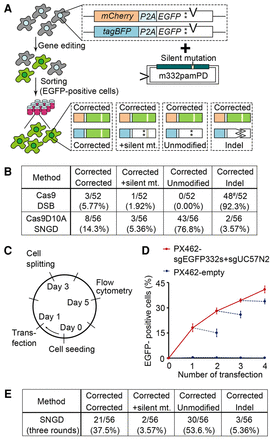
Sequence analysis of the reporter gene after the SNGD-mediated gene editing. (A) Diagram of the nucleotide substitution assay using the bi-EGFPcC>G reporter cells. (*) c.321C>G mutation. (B) Summary of the sequence analysis of the EGFPcC>G reporter in the EGFP-positive bi-EGFPcC>G reporter single-cell clones. The clones were classified into four types based on the sequencing results: (1) Both reporters were corrected as designed; (2) one copy was corrected and the other incorporated the silent mutation of the repair template, but the c.321C>G mutation was not corrected; (3) one copy was corrected and the other was not modified; and (4) one copy was corrected and the other resulted in indel (also see A): (#) One clone contained two nucleotide substitutions near the DSB site without an indel. (C–E) The SNGD process was repeated. (C) The schedule of the SNGD cycle is shown. (D) Gene-editing efficiency as measured by flow cytometry is shown (mean ± SD, N = 3). (Solid line) SNGD; (dashed line) no nick. (E) A summary of the sequence analysis of the EGFPcC>G reporter in the EGFP-positive bi-EGFPcC>G reporter single-cell clones after three consecutive rounds of the SNGD process.











