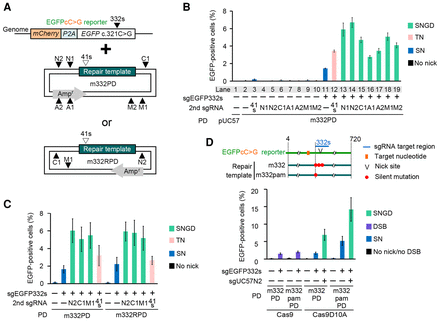
Nucleotide substitution by a combination of single nicks in the reporter gene and backbone of the donor plasmid (SNGD). (A) Schematic of the nucleotide substitution reporter assay by SNGD. sgRNA target sites are indicated by triangles. (B,C) Nucleotide substitution efficiency as measured by flow cytometry (mean ± SD, N = 3 [B] or N = 4 [C]) is presented in each panel. A nick in the EGFPcC>G reporter was introduced at the sgEGFP332s site. A nick in the backbone of the donor plasmid was introduced at one of the sgUC57 sites. m332PD (B) or m332RPD (C) was used as the donor. (D) Efficiency of the SNGD-mediated or DSB-mediated nucleotide substitution as measured by flow cytometry (mean ± SD, N = 3) is presented.











