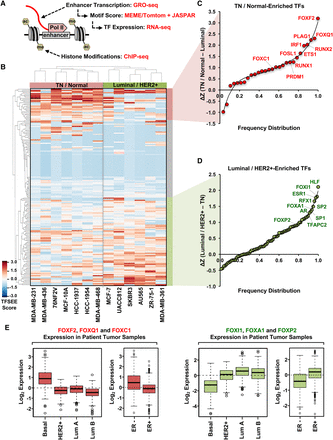
A functional score of enhancer elements identifies subtype-specific enhancers and their cognate TFs that drive subtype-specific gene expression in breast cancer cells. (A) Diagram of the data used for determining the Total Functional Score of Enhancer Elements (TFSEE) across breast cancer cell lines. TFSEE simultaneously identifies putative subtype-specific enhancers and their cognate TFs by integrating the magnitude of enhancer transcription (GRO-seq), TF mRNA expression levels (RNA-seq), TF motif P-values (MEME/Tomtom), and enrichment of H3K4me1 and H3K27ac (ChIP-seq). This analysis yields the location, activity level, and predicted TFs at each enhancer in all breast cancer cells. (B) Unsupervised hierarchical clustering of cell line–normalized TFSEE scores shown in a heatmap representation. Two major clades arise from this analysis, highlighting key TFs for TN/Normal subtypes versus Luminal/HER2+ subtypes. (C,D) Rank order frequency distribution of TFs enriched in the TN/Normal-like clade (C) and the Luminal/HER2+ clade (D) identified using TFSEE. The top TFs in each clade are noted. (E) Box plots of expression values for members of the Forkhead box family of TFs in patient breast tumor samples, confirming the differential enrichment of these TFs in the TN/Normal-Like versus the Luminal/HER2+ clades shown in C and D. Observed differences are significant as determined by an ANOVA comparison of the means (P-value < 0.00001).











