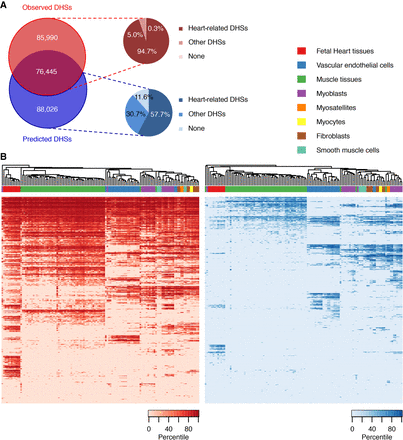
Learned sequence features predict additional CREs. (A) Venn diagram of observed and predicted DHSs and their proportions in noncardiac cell types. (B) Heat map of DHS signal intensities in heart-related cell types and tissues at observed (left) and predicted (right) DHSs. Randomly sampled 5000 regions from observed and predicted DHSs were used. Observed DHSs are mostly located in ubiquitously open chromatin regions in cardiac-relevant cells and tissues, while predicted DHSs show greater cell-type specificity.











