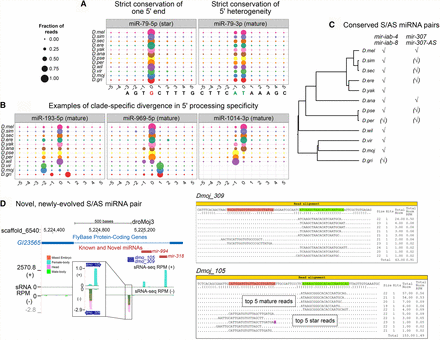
Shifted processing and alternate biogenesis pathways of Drosophila miRNAs. (A,B) Consistency in 5′ end processing for conserved miRNAs. (A) sRNA read alignment for mir-79, represented compactly in these bubble plots, show two miR sequences with unique 5′ ends. These represent two seed-distinct iso-miRs that are both produced in several Drosophila species. Position 0 represents the proportion of reads that begin with the base of the most abundant 5′ arm sequence at either the 5′ strand (miR*) and 3′ strand (miR) for all 12 Drosophila genomes. Proportions shown at positions less than or greater than 0 represent proportion of reads with shifted processing. For mir-79, two iso-miRs are produced in similar proportions. (B) Panels of bubble plots depict the heterogeneity of 5′ end processing for the miR sequence of other conserved miRNAs and mirtrons. Greater than 4 alternate iso-miR-193 sequences in D. melanogaster were noted previously. This heterogeneity is preserved in the genomes of the other Drosophilids, and conserved, dominant iso-miRs is not apparent. We identified clade-specific iso-miRs for one canonical miRNA (mir-969) and one mirtron (mir-1014). Specifically, two unique iso-miR-969 sequences are each preferentially abundant in the Sophophora group and Drosophila group species, respectively, and for mir-1014, the melanogaster group species produces one iso-miR-1014 sequence that is distinct from the dominant iso-miR of other Sophophorans. (C) mir-iab-4/8 and mir-307/mir-307-as are the only two reasonably conserved miRNAs with sense and antisense transcription and processing based upon our genus-wide data. (D) We identified dozens of recently evolved antisense miRNAs; shown is the example of dmo_105/dmo_309 that arose adjacent to the conserved mir-994/318 cluster.











