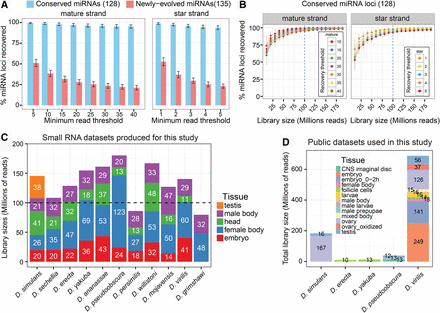
Summary of Drosophila species small RNA sequencing data and analysis of sequencing depth sufficiency. (A) The recovery rate of known D. melanogaster miRNAs using sets of 100 million total reads sampled randomly from D. melanogaster head, mixed embryo, male-body, and female-body public libraries. These libraries mimic those we sought to create for the 11 other Drosophila genomes. Bars represent the fraction of conserved or newly evolved D. melanogaster miRNAs recovered at various miRNA and miRNA* minimum read thresholds, and error bars represent the standard error of the recovery rate across 100 independent samples of 100 million reads. (B) Saturation curve of miRNA (mature and star) strand recovery at varying minimum read depth cutoffs. Based upon these results, we sought to acquire approximately 100 million reads per species. (C) Summary statistics of the actual Drosophila species small RNA libraries across tissues generated for this study. (D) Summary of publicly available Drosophila species small RNA libraries utilized in this study.











