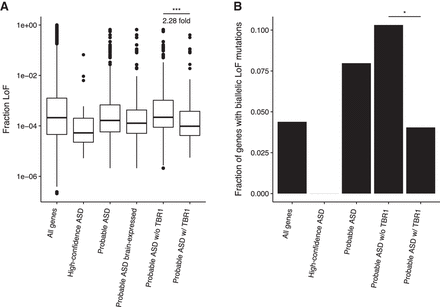
Probable ASD genes that are TBR1 targets are more depleted for ExAC LoF mutations and biallelic LoF mutations in Icelanders. (A) Box plots depicting the distributions of fraction LoF scores for each gene from the ExAC reference population (y-axis) for merged ASD gene lists (x-axis). Probable ASD genes with adjacent TBR1 ChIP-seq peaks in the developing cortex have lower fraction LoF scores than those without an adjacent TBR1 peak. A pseudocount of one LoF allele for 121,412 sampled alleles, the maximum number sampled at any locus, was included for each gene for visualization purposes. Significance was determined using the one-sided two-sample Wilcoxon test. (B) The fraction of genes in each gene list with biallelic mutations in a study of Icelandic individuals (Sulem et al. 2015). Significance was determined using the one-sided Fisher's exact test. (*) P-value < 0.05; (***) P-value < 0.001.











