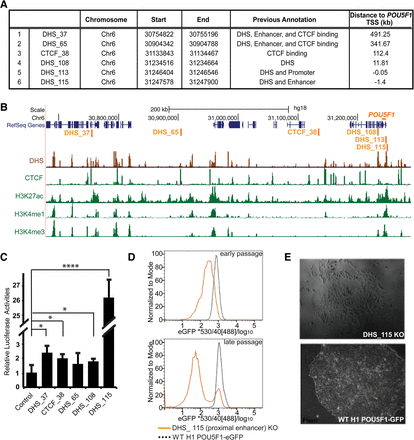
Characterization of cis-regulatory elements identified near POU5F1. (A) A list of regulatory elements regulating POU5F1 transcription identified from the lentiCRISPR screening with coordinates (hg18). Each element was identified by at least two distinct sgRNAs enriched by twofold or more in the eGFP− population in at least three out of the four independent screenings. (B) Genome browser snapshot shows the location and epigenetic environment for each element in H1 hESCs. (C) Reporter assays for the six identified cis-regulatory elements. H1 hESC cells were transfected with various luciferase reporter plasmid as indicated. Two days post transfection, cells were lysed and subjected to luciferase reporter assays. All tested elements are cloned into the control reporter plasmid containing the 360-bp POU5F1 minimal promoter sequence driving reporter gene expression. DHS_115 exhibits the highest enhancer activities ([****] P < 10−6), while the other four elements show either minimal (DHS_37, CTCF_28, and DHS_108 with [*] P < 0.05) or no enhancer activities (DHS_65 with P = 0.27) compared with the control reporter plasmid containing POU5F1 promoter only. (D) FACS analysis showed that the DHS_115 biallelic mutant hESCs (orange line) gradually lose POU5F1-eGFP expression in culture over time compared with parental H1 POU5F1-eGFP line (dash line). (E) Phase images of hESCs with biallelic mutations at DHS_115 locus (top) and wild-type hESCs (bottom). Deletions of DHS_115 sequences lead hESCs to lose pluripotency and become differentiated in the culture (top).











