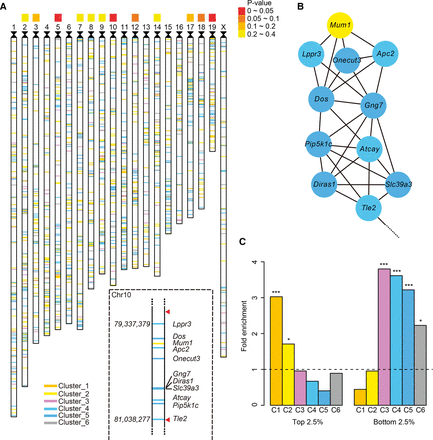
Role of chromatin remodeling in neurogenesis. (A) The ideogram of all genes curated by the image analysis pipeline. No genes on Chr Y were detected. Other chromosomes in mouse are acrocentric; centromeres are indicated with black triangles. The color-coded panel indicates P-values for chromosomal clustering of genes from anti-correlated modules. (Dashed box) A zoom-in on the 1.7-Mb Chr 10 domain, with the genome coordinates of TSS on the left and red triangles indicating the Hi-C domain boundary. (B) The Hi-C interaction network between genes in the 1.7-Mb Chr 10 domain. (C) The fold enrichment of genes with the top or bottom 2.5% neural progenitor cells (NPCs) versus neuron H3K4me3/H3K27me3 ratios in each cluster (labeled C1–C6), estimated based on 100,000 permutation background (dashed line). Asterisks indicate empirical P-values by permutation (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. The colors of ideogram lines, network nodes, and bar plots denote different expression clusters, respectively: orange, Cluster 1; yellow, Cluster 2; purple, Cluster 3; cyan, Cluster 4; blue, Cluster 5; gray, Cluster 6.











