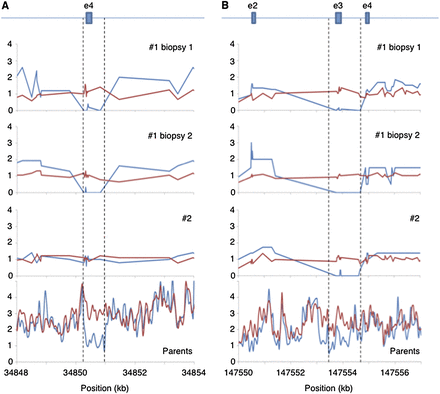
Detection of heterozygous deletions of small exons. LFR haplotype information can be used to separate coverage for each allele. Normalized coverage from each LFR haplotype for embryo #1 biopsies 1 and 2 and embryo #2, as well as 50-bp read coverage windows for both parents, were plotted (blue indicates father; red, mother). (A) A heterozygous deletion of ∼500 bp in the gene TTC23L removing all of exon 4 and part of the intron on either side in both biopsies of embryo #1 and the father was detected. (B) A heterozygous deletion of ∼1000 bp in the gene SPINK14 removing all of exon 3 and parts of the intron on either side was identified in all three biopsies. Coverage for the parents is more difficult to interpret in this region, but it appears that again the father has less coverage.











