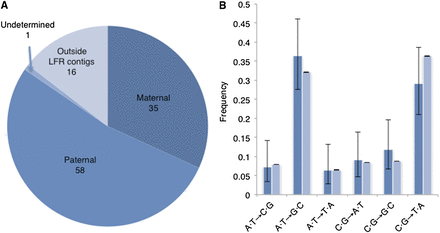
Characteristics of de novo SNVs in embryo #1. After filtering, 110 putative de novo SNVs (including about 10 errors) were identified in embryo #1. (A) Phasing enabled the parent of origin to be determined for 93 of the de novo SNVs. Similar to previous studies, almost twice as many de novo SNVs came from the father as compared to the mother. (B) Specific nucleotide changes for de novo (dark blue) and inherited (light blue) were plotted by frequency. Frequencies of nucleotide changes were similar between de novo and inherited, as would be expected for true de novo SNVs. 95% confidence interval error bars were computed using a one sample proportions test, allowing for Yates’ continuity correction using R software (R Core Team 2014). The error bars suggest that the small differences observed between de novo and inherited are insignificant.











