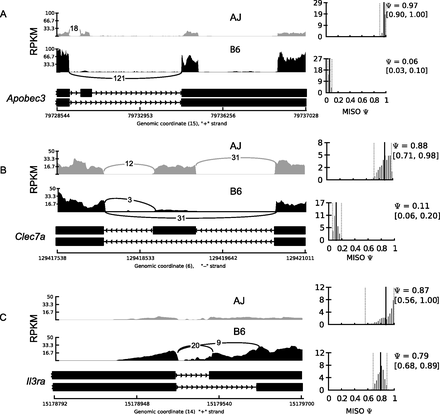
Isoform usage varies between laboratory mouse strains. We observed differential isoform usage (reflected by the different PSI values) of Apobec3 (A), Clec7a (B), and Il3ra (C) between the different mouse strains. Shown are the percent-splice-in (PSI, ψ) for the longest isoform for each gene, and the lower and upper 95% confidence intervals (CIs) (in parentheses). Also shown is the total number of reads supporting each splice junction. Due to the low, almost absent, expression of the alternative Il3ra isoform in the AJ, which is consistent with a previous observation (Ichihara et al. 1995), there are no reads supporting the alternative splice junction, which is also the reason for the large CI. The alternative isoforms for both Apobec3 and Clec7a are due to a skipped exon, while the Ilr3a alternative isoform is due to an alternative 3′ splice site (alternative acceptor).











