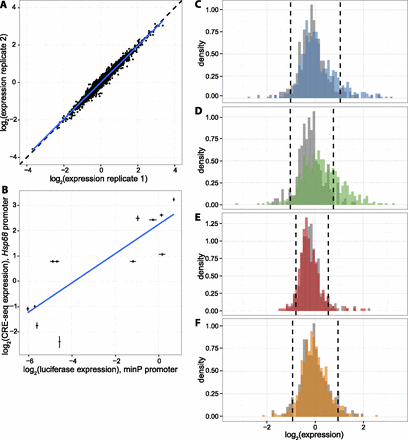
Reproducible expression measurements show differences in expression by segmentation class. (A) Representative scatterplot showing expression of each CRE in two biological replicates (R2 = 0.95, range of R2 between all replicates: 0.95–0.97). Dashed black line is line of equality and blue line is best fit. (B) Correlation between CRE-seq and luciferase assays. Expression driven by 12 CREs was measured in individual luciferase assay (upstream of minP promoter, x-axis) and batch CRE-seq assay (upstream of Hsp68 promoter, y-axis). Luciferase expression is normalized to the Renilla transfection control, and CRE-seq expression is normalized to the basal promoter alone. Error bars represent the standard error of the mean. Blue line is best fit. R2 = 0.70. (C–F) Histograms of genomic CRE expression measurements in K562 cells. Each class is compared to scrambled controls with equivalent GC and dinucleotide content (gray). Dashed lines are the fifth and 95th percentiles of the scrambled distributions. (C) K562 Enhancer class (blue), (D) K562 Weak Enhancer class (green), (E) K562 Repressed class (red), (F) H1-hESC Enhancer class (orange).











