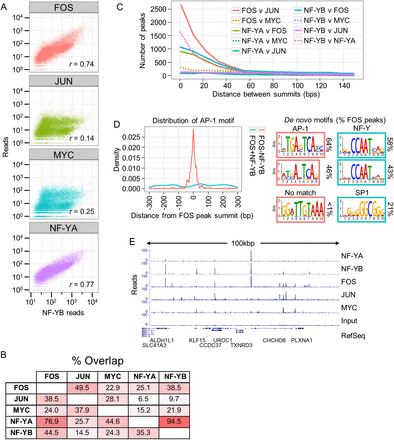
NF-Y and FOS are closely coassociated at loci that lack JUN and the AP-1 motif. (A) Correlation between ChIP-seq read counts at NF-YB peak summits, within a window of ±500 bp, between NF-YB and NF-YA, FOS, JUN, or MYC in K562 cells. (B) Values represent the percentage of peak populations (left row) directly overlapping the peak population of a second factor (top column). All binding sites are called at a P-value < 10−9. FOS (n = 14404); JUN (n = 18480); MYC (n = 13693); NF-YA (n = 4726); NF-YB (n = 12655). (C) The number of ChIP-seq peaks at the indicated distance between adjacent peak summits is plotted. All peaks were called at a 10−9 P-value threshold in K562. (D) The top 1000 K562 FOS ChIP-seq sites, as ranked by site P-value, that directly overlap an NF-YB site (“FOS+NF-YB”) and the top 1000 that do not overlap an NF-YB site (10−5 P-value site list, “FOS-NF-YB”) are assayed for the distribution of the AP-1 motif in relation to the FOS peak summit centered at 0 bp. Plotted is the Gaussian kernel density estimate of the AP-1 motif using a bandwidth of 0.5 of the standard deviation of the smoothing kernel. The top three motifs discovered de novo from each FOS peak set, as above, are depicted with the percentage of FOS peaks containing a match to that motif indicated. (E) Representative view of a locus on chromosome 3 of the K562 ChIP-seq read counts from NF-YA, NF-YB, FOS, JUN, and MYC ChIPs, with an input control.











