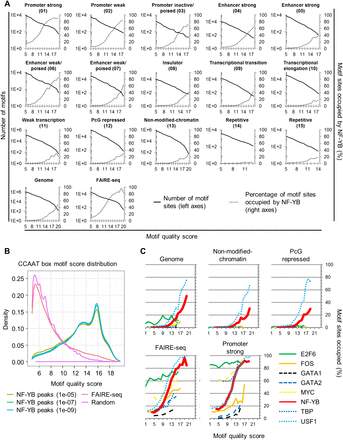
NF-YB can occupy its motif in closed chromatin. (A) The percentage of genome-wide computationally discovered CCAAT motifs within each chromatin state, FAIRE-seq regions or the entire genome, that directly overlap NF-YB K562 sites plotted as a function of CCAAT motif quality (right axes). Also shown are the numbers of discovered CCAAT motifs as a function of quality (left axes). Numbering is derived from Ernst et al. (2011) and kept for consistency. (B) Distribution of CCAAT motif quality scores under NF-YB K562 peaks, called at three different P-values, a random genomic background sample set of 400 k 500-bp regions and K562 FAIRE-seq regions. (C) Similar to A, except motifs of different TFs are plotted as a function of motif quality. Only a subset of TFs is shown; see Supplemental Figure 13 for all TFs analyzed.











