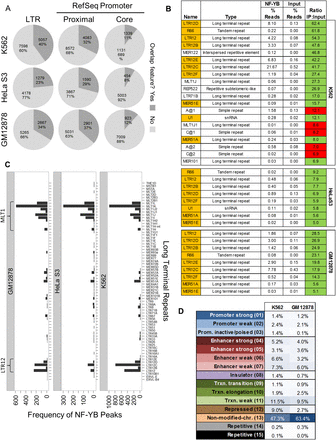
NF-YB binds extensively to long terminal repeats. (A) The percentage of all K562 NF-YB peak summits that occupy the indicated feature. Core and proximal promoters are defined as −250:+50 bp and −2500:+500 bp from the TSS of RefSeq promoters, respectively. (B) Mapping of ChIP-seq reads from K562, GM12878, and HeLa S3 to Repbase consensus sequences showing an abundance of NF-Y ChIP-seq reads mapping to repetitive elements. Ratios reflect the enrichment of reads in the NF-YB ChIP sample as compared with input. Only Repbase entries with a read ratio ≥5 are shown. Orange shading indicates enriched repeats present in all cell lines. Green and red shading indicate the presence and absence, respectively, of a CCAAT motif match at P-value < 10−4 in the consensus sequence. (C) Frequency of overlap between NF-YB peak summits and the genomic locations of LTR families. Only LTR elements that overlap at least one NF-YB summit in each cell line are shown. The two most highly overlapping repeat families are indicated, LTR12 and MLTJ1. (D) Distribution of NF-YB bound LTRs from K562 and GM12878 at chromatin states. No chromatin state map is available for HeLa S3.











