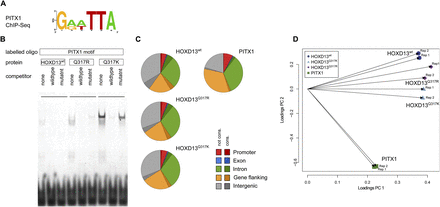
The HOXD13Q317K mutant binds the PITX1 binding site in vitro, and the ChIP-seq profile shifts toward PITX1. (A) The primary PITX1 motif identified from ChIP-seq. (B) In vitro binding of wild-type and mutant HOXD13 homeodomains to the PITX1 motif. EMSAs were performed as in Fig. 1C using oligonucleotides containing a binding sequence for PITX1 (GGATTA). HOXD13Q317K specifically binds the PITX1 motif, whereas it is not bound by HOXD13wt or HOXD13Q317R. (C) Genomic distribution of ChIP-seq peaks. The genome was divided into promoter (5 kb upstream of, 2 kb downstream from TSS), exon, intron, gene flanking (20 kb upstream, 20 kb downstream), and intergenic regions, each separated in conserved and nonconserved regions. (D) Principle component analysis for the coverage profiles of the uniquely mapped reads for all eight data sets (two biological replicates each for HOXD13wt, HOXD13Q317K, HOXD13Q317R, and PITX1).











