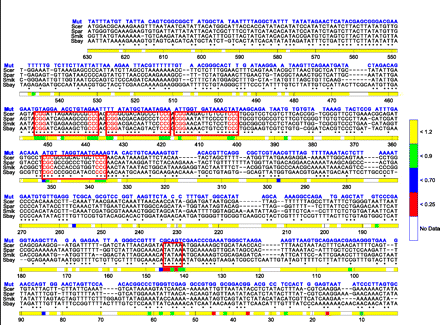
Multiple sequence alignments among four yeast species in the GAL1-10 regulatory region and gene expression of single-nucleotide mutated strains. The sequence alignment is shown according to Kellis et al. (2003). (Axis) The position from −1 to −630 before the GAL1 start codon. (*) Conserved positions among four species. (Sequences in blue) The mutated nucleotides. Validated Gal4 and TATA binding sites are boxed in red. Gene expression variations for a single nucleotide mutation are indicated with different colors along the position axis in the expression bar. The color bar indicates different expression levels (see color map). (Scer) S. cerevisiae; (Spar) S. paradoxus; (Smik) S. mikatae; (Sbay) S. bayanus; (Mut) mutated nucleotide.











