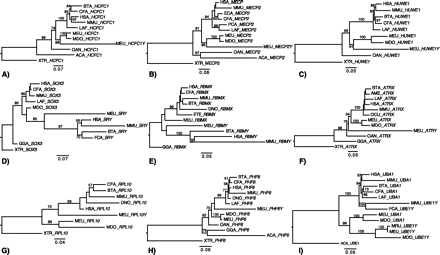
Maximum likelihood trees of marsupial XY gene pairs. Bootstrap values >50% are shown. These analyses indicate that (A) HCFC1X/Y, (B) MECP2X/Y, and (C) HUWE1X/Y were members of the ancestral therian PAR at the time of the marsupial/eutherian split; the (D) SOX3/SRY, (E) RBMX/Y, and (F) ATRX/Y gene pairs were isolated from each other before the marsupial/eutherian split; and (G) RPL10X/Y, (H) PHF6X/Y, and (I) UBA1/UBE1Y have a more complex evolutionary history (see text). Species code: (HSA) Human; (MMU) mouse; (OCU) rabbit; (CFA) dog; (FCA) cat; (BTA) cow; (ECA) horse; (AME) panda; (DNO) armadillo; (LAF) elephant; (PCA) hyrax; (ETE) tenrec; (MDO) Monodelphis domestica; (MEU) tammar wallaby; (MRU) Macropus rufus; (OAN) platypus; (GGA) chicken; (ACA) anolis; (XTR) Xenopus tropicalis. Trees reflect outgroup rooting.











