Evolutionary history of novel genes on the tammar wallaby Y chromosome: Implications for sex chromosome evolution
- Veronica J. Murtagh1,2,
- Denis O'Meally1,3,
- Natasha Sankovic1,2,4,
- Margaret L. Delbridge1,2,
- Yoko Kuroki5,
- Jeffrey L. Boore6,7,
- Atsushi Toyoda8,
- Kristen S. Jordan1,
- Andrew J. Pask2,4,9,
- Marilyn B. Renfree2,4,
- Asao Fujiyama8,10,
- Jennifer A. Marshall Graves1,2,4 and
- Paul D. Waters1,2,11
- 1Evolution, Ecology and Genetics, Research School of Biology, The Australian National University, Canberra, ACT 2601, Australia;
- 2ARC Centre of Excellence for Kangaroo Genomics, The University of Melbourne, Victoria 3010, Australia;
- 3Institute for Applied Ecology, University of Canberra, ACT 2601, Australia;
- 4Department of Zoology, The University of Melbourne, Victoria 3010, Australia;
- 5RIKEN Research Center for Allergy and Immunology, Immunogenomics, Yokohama, 230-0045, Japan;
- 6Genome Project Solutions, Hercules, California 94547, USA;
- 7DOE Joint Genome Institute, Walnut Creek, California 94598 USA;
- 8National Institute of Genetics, Mishima, 411-8540, Japan;
- 9Department of Molecular and Cellular Biology, University of Connecticut, Storrs, Connecticut 06260, USA;
- 10National Institute of Informatics, Tokyo, 101-8430, Japan
Abstract
We report here the isolation and sequencing of 10 Y-specific tammar wallaby (Macropus eugenii) BAC clones, revealing five hitherto undescribed tammar wallaby Y genes (in addition to the five genes already described) and several pseudogenes. Some genes on the wallaby Y display testis-specific expression, but most have low widespread expression. All have partners on the tammar X, along with homologs on the human X. Nonsynonymous and synonymous substitution ratios for nine of the tammar XY gene pairs indicate that they are each under purifying selection. All 10 were also identified as being on the Y in Tasmanian devil (Sarcophilus harrisii; a distantly related Australian marsupial); however, seven have been lost from the human Y. Maximum likelihood phylogenetic analyses of the wallaby YX genes, with respective homologs from other vertebrate representatives, revealed that three marsupial Y genes (HCFC1X/Y, MECP2X/Y, and HUWE1X/Y) were members of the ancestral therian pseudoautosomal region (PAR) at the time of the marsupial/eutherian split; three XY pairs (SOX3/SRY, RBMX/Y, and ATRX/Y) were isolated from each other before the marsupial/eutherian split, and the remaining three (RPL10X/Y, PHF6X/Y, and UBA1/UBE1Y) have a more complex evolutionary history. Thus, the small marsupial Y chromosome is surprisingly rich in ancient genes that are retained in at least Australian marsupials and evolved from testis–brain expressed genes on the X.
Most mammals have an XX female/XY male sex chromosome system, in which maleness is determined by a single dominant gene (SRY) on a poorly conserved Y chromosome. The euchromatic region of the human male-specific region of the Y (MSY) is relatively small (∼25 Mb) and contains 172 transcription units that code for 27 distinct proteins, many of which are testis specific and involved in spermatogenesis (Lahn and Page 1997; Skaletsky et al. 2003). At least two genes were acquired via retrotransposition/transposition from autosomes (Saxena et al. 1996; Lahn and Page 1999b), and at least 20 have partners on the X (for review, see Waters et al. 2007) from which they evolved (Graves 1995).
In contrast, the X chromosome is well conserved between species (Raudsepp et al. 2004a; Rodriguez Delgado et al. 2009). The human X is 155Mb (Ross et al. 2005) and bears 1669 genes (NCBI database: http://www.ncbi.nlm.nih.gov). Along with X/Y shared genes, the X shares homology with the Y chromosome in two small terminal pseudo-autosomal regions, the larger of which is critical to the proper pairing and segregation of the X and Y at male meiosis.
Although the X and Y are morphologically and functionally distinct, they evolved from an autosomal pair (Ohno 1967). After the proto-Y acquired a testis-determining factor (TDF), male beneficial genes accumulated nearby, and recombination in the region was suppressed so that these genes were inherited together only in males. In the absence of recombination, the Y degraded because of inefficient selection, genetic drift, and increased variation (Charlesworth 1991). On the human Y, the 20 genes with an X homolog are all that survived this degradation process.
The process of degradation has not been uniform. Both the X and Y are composed of ancient and added regions, defined by comparing sex chromosomes between eutherian and marsupial mammals. The ancient region (XCR/YCR) is conserved on the X in both groups, but is autosomal in monotremes (Veyrunes et al. 2008), implying that it started differentiating between 148 and 166 million years ago (MYA) (Bininda-Emonds et al. 2007), and that it contributes only four genes to the human Y. The added region (XAR/YAR) is autosomal in marsupials, but on the sex chromosomes in all eutherian mammals, so it fused to the X and Y between 148 and 105 MYA (Graves 1995, 2006). It is the source of nearly all of the human Y genes (Waters et al. 2001). The human X is further subdivided (by comparing synonymous substitution rates between XY gene pairs) into four evolutionary strata (Strata 1 and Strata 2, XCR; Strata 3 and Strata 4, XAR), each representing an event (perhaps inversions on the Y) that suppressed recombination with the Y (Lahn and Page 1999a; Skaletsky et al. 2003).
Much knowledge of the mammal Y degradation process comes from the detailed study of human and chimpanzee Y chromosomes, which show significant divergence for such closely related species (Skaletsky et al. 2003; Hughes et al. 2005, 2010; Kuroki et al. 2006). Y chromosome studies in mouse, cat, and horse (Raudsepp et al. 2004b; Toure et al. 2005; Pearks Wilkerson et al. 2008) have revealed even greater variability of the Y gene content in more distantly related eutherian mammals and highlight the remarkable nature of linage-specific Y chromosome gene loss, retention, and gain.
Since marsupial and eutherian mammals diverged from each other early in the history of X–Y differentiation, the marsupial X and Y represent a largely independent example of mammalian sex chromosome differentiation. It is therefore of great interest to examine a marsupial Y chromosome to better understand the general principles by which mammalian Y chromosomes (indeed, all sex-limited chromosomes) evolved. The marsupial X and Y lack a pseudoautosomal region (Sharp 1982), so are terminally differentiated and pair at meiosis by a completely different mechanism from that of eutherian mammals (Page et al. 2005, 2006). The Y chromosome of the model kangaroo (tammar wallaby, Macropus eugenii), for which a genome assembly was recently released (Renfree et al. 2011), contains orthologs (separated by speciation) of three eutherian Y genes (SRY, RBMY, and KDM5D) (Foster et al. 1992; Delbridge et al. 1999; Waters et al. 2001), along with the marsupial-specific ATRY (Pask et al. 2000). ATRY is also on the Y in the gray short-tailed opossum (Carvalho-Silva et al. 2004), but was lost from the Y chromosome in a eutherian mammal ancestor. The possibility remains that the repetitive long arm of the tammar wallaby Y, which was acquired in the macropod ancestor and consists of degenerate NOR sequence (Toder et al. 1997), could bear retrotransposed genes that have been amplified and specialized.
We report here the isolation and sequencing of 10 Y-specific tammar wallaby BAC clones that contained several novel genes, all with partners on the tammar X, and all conserved on the Y in a second Australian marsupial. Nonsynonymous/synonymous substitution ratios between nine XY gene pairs indicated that the Y genes are under purifying selection. Maximum likelihood phylogenetic analyses revealed a complex evolutionary history for some of the Y genes, and suggest that at least three marsupial-specific Y genes were members of the ancestral therian PAR.
Results
We used flow-sorted tammar Y chromosome DNA and Y-specific probes for three previously identified Y genes (RBMY, SRY, ATRY) to isolate Y chromosome-specific BAC clones. Physical locations were assessed by fluorescence in situ hybridization (FISH). We sequenced 10 Y BACs, identified five novel X–Y shared genes, and studied their expression. We also investigated whether genes on the tammar Y chromosome were located on the Tasmanian devil Y chromosome by searching for evidence of Y chromosome ESTs in the Tasmanian devil transcriptome (Murchison et al. 2010).
BAC library screening
Y chromosome-specific BAC clones were isolated from three different tammar male-derived BAC libraries (ME_VIA, ME_Kba, and MEB10 (Table 1). BACs were isolated using probes for RBMY, SRY, and ATRY, which were generated by PCR, radioactively labeled, and used to screen filters for the ME_VIA BAC library. This yielded three BAC clones (ME_VIA-53A23 - ATRY, ME_VIA-80O22 – RBMY, and ME_VIA-112D12 - SRY). The MEB1 BAC library was screened for clones containing SRY and RBMY by two-step three-dimensional (3D) PCR screening. This strategy yielded two clones: one (MEB1-052D08) containing SRY and a second (MEB1-321E03) containing RBMY. Five other clones isolated from ME_VIA and ME_KBa (with Y chromosome mircodissected paints) (Sankovic et al. 2006) were also chosen for sequencing.
BAC clones sequenced in this study
BAC mapping
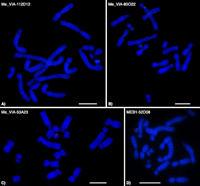
BAC clones were mapped by FISH. About half produced signals on the Y chromosome, but most of these showed repetitive Y-specific signals (Supplemental Fig. 1; Sankovic et al. 2006). The BAC clones identified above produced punctate signals on the Y chromosome (Fig. 1), suggesting that they contain a low copy-number region of the tammar Y; these were chosen as top priority for sequencing.
Sequence analysis
The five ME_VIA BACs were sequenced and assembled at the Joint Genome Institute. The MEB1 and ME_KBa clones were sequenced and assembled by the Comparative Genomics Laboratory at the National Institute of Genetics (NIG). BAC sequences are available in the NCBI nucleotide database (Table 1).
We used GenScan (http://genes.mit.edu/GENSCAN.html) to predict the exon–intron structures of genes in all these BAC clones. Predicted protein sequences were identified by querying the translated nucleotide nonredundant database (TBLASTN, NCBI). The entire sequences of the BAC clones were used to search both the opossum and human genomes using default parameters for the discontiguous megablast algorithm. These methods identified a total of five new marsupial Y-borne genes (RPL10, MECP2, HCFC1, HUWE1, and PHF6) (Table 1), all with an X partner. None of these new tammar Y genes have been identified on the Y in a eutherian mammal.
We found evidence of two apparently ancient retrotransposed pseudogenes (PYGM on human chromosome 11; and SLC3A1 on human chromosome 2) and an X degenerate pseudogene (FAM122C). Low amino acid homology (40%–60%) to PYGM, SLC3A1, and FAM122C was only detected using tblastx searches (never nucleotide blast searches), and they could not be aligned to their progenitor genes. GenScan predicted no corresponding open reading frames, and no further analyses were conducted on these pseudogenes. We also found evidence of a recently retrotransposed gene (SPATA2Y), which shares high-sequence identity (95%) with its autosomal copy (SPATA2 on human chromosome 20). However, SPATA2Y is truncated after 100 amino acids (which represents only the first 20% of the full protein) relative to both the human and wallaby SPATA2, which together with a Ka/Ks ratio of close to 1 for wallaby SPATA2 and SPATA2Y (see below) indicates that SPATA2Y is not under selective pressure and is a pseudogene.
The repetitive content of the BAC clone sequences was analyzed using RepeatMasker (http://repeatmasker.org) (Table 2). Clones that mapped in the vicinity of SRY (MEKBa-112D12 and MEB1-052D08) had a much lower content of total interspersed repeats compared with the genome average for opossum (52.2%) and human (45.5%) (Venter et al. 2001; Mikkelsen et al. 2007). In contrast, the total repeat content in the ATRY and RBMY regions was similar to the opossum and human genome average. Repeat elements within the Y BACs were predominantly LINEs, with a lower than genome average number of SINEs, ERVs, and DNA transposons. Five of the BAC clones bore no detectable repeats, probably because repeats in these Y BACs are not represented in the RepeatMasker database. They were therefore excluded from calculations of total repeat content of all sequenced BACs (Table 2).
Repeat content of BAC clones in this study
X-borne gene copies of tammar Y genes
The physical locations of all X-borne copies were assessed to confirm that these genes were located on the X chromosome, and to ascertain their relative positions on the X. X-borne copies of all 10 Y genes were identified in the Ensembl wallaby assembly (http://www.ensembl.org/Macropus_eugenii/Info/Index). Large-insert genomic clones containing eight of these genes (ATRX, HUWE1X, JARID1C, MECP2X, PHF6X, RBMX, SOX3, and UBA1) were already isolated and physically mapped (Deakin et al. 2008). Predicted transcripts from the two previously unmapped genes (HCFC1X and RPL10X) were used to query end sequences from a tammar X chromosome-enriched fosmid library (MEFX) using megablast. We used PCR to confirm the identified fosmids containing HCFC1X (MEFX-044I17) and RPL10X (MEFX-003A16). Using two-color FISH, we determined that HCFC1X and RPL10X map to Xq1 in wallaby, and using a BAC bearing MECP2X (MEKBa-143H17) we confirmed that the gene order from the centromere is RPL10X–MECP2X–HCFC1X.
Nucleotide similarity of X and Y homologs
Synonymous (Ks) and nonsynonymous (Ka) substitution rates were calculated for each wallaby X–Y shared gene (for which sequence data were available). Rates were also calculated for the wallaby X and human X orthologs. Ka/Ks ratios of <1 for all marsupial X–Y gene pairs (Table 3) indicate purifying selection. There were no clear groupings of Ks values to suggest well-defined evolutionary strata on the tammar wallaby X (see Supplemental Table 1). To better elucidate the evolutionary history of tammar wallaby Y genes, phylogenetic analyses were undertaken.
Alignment details of genes in this study and Ka/Ks ratios
Phylogenetic analyses of XY gene pairs
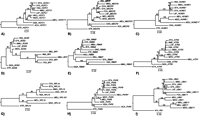
The tammar wallaby HCFC1Y, MECP2Y, and HUWE1Y cluster with the marsupial X homologs (Fig. 2), suggesting that they diverged from their X partners after the marsupial/eutherian split. The therian X copies of SOX3/SRY, RBMX/Y, and ATRX/Y cluster together (although there is poor bootstrap support), suggesting that the X–Y gametologues were isolated from each other before the marsupial/eutherian split. RPL10X/Y (and perhaps PHF6X/Y, although there is poor support) displayed an unusual topology (Fig. 2), with the marsupial Y copy clustering with the eutherian X copies, suggesting a more complex evolutionary history. Finally, the marsupial UBE1Y clustered with the marsupial UBA1, and the eutherian UBE1Y clustered with the eutherian UBA1 (Fig. 2).
Maximum likelihood trees of marsupial XY gene pairs. Bootstrap values >50% are shown. These analyses indicate that (A) HCFC1X/Y, (B) MECP2X/Y, and (C) HUWE1X/Y were members of the ancestral therian PAR at the time of the marsupial/eutherian split; the (D) SOX3/SRY, (E) RBMX/Y, and (F) ATRX/Y gene pairs were isolated from each other before the marsupial/eutherian split; and (G) RPL10X/Y, (H) PHF6X/Y, and (I) UBA1/UBE1Y have a more complex evolutionary history (see text). Species code: (HSA) Human; (MMU) mouse; (OCU) rabbit; (CFA) dog; (FCA) cat; (BTA) cow; (ECA) horse; (AME) panda; (DNO) armadillo; (LAF) elephant; (PCA) hyrax; (ETE) tenrec; (MDO) Monodelphis domestica; (MEU) tammar wallaby; (MRU) Macropus rufus; (OAN) platypus; (GGA) chicken; (ACA) anolis; (XTR) Xenopus tropicalis. Trees reflect outgroup rooting.
Expression of X and Y gene pairs
The expression patterns of tammar X and Y alleles of each gene were examined by quantitative PCR on RNA extracted from male tammar wallaby tissues (brain cortex, kidney, liver, lung, spleen, testis, fibroblast) plus ovary. We compared the expression of the X and Y gametologues in these tissues for eight marsupial X–Y gene pairs (HCFC1X/Y, RPL10X/Y, PHF6X/Y, RBMX/Y, ATRX/Y, HUWE1X/Y, UBA1/UBE1Y, and MECP2X/Y). The detection of a male-specific UBA1 transcript provided the first direct evidence of a tammar Y-borne homolog.
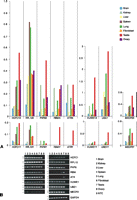
Expression patterns of RBMX/Y and ATRX/Y in tammar have been explored previously by nonquantitative reverse-transcription PCR (RT–PCR) (Delbridge et al. 1997; Pask et al. 2000). The X copies of both of these genes were widely expressed, whereas the Y copies were testis specific. Our quantitative-PCR (qPCR) results for these two genes were consistent with these previous studies, although very low-level expression of RBMY was detected in kidney, lung, and spleen (Fig. 3).
Expression pattern of tammar Y genes and their X homologs, assessed by (A) qPCR and (B) endpoint PCR in different tissues (NTC: no template control). All qPCR results were normalized to the autosomal gene GAPDH.
The X copies of UBA1/UBE1Y, PHF6X/Y, HUWE1X/Y, HCFC1X/Y, MECP2X/Y, and RPL10X/Y (Fig. 3) were all widely expressed. However, in contrast to RBMY and ATRY, wide expression was also detected for the Y gametologue. For all but one of these gene pairs (MECP2X/Y), the Y copy was expressed at a lower level than the X copy in all tissues (Fig. 3). For MECP2X/Y, expression of the X and Y copies were nearly equivalent (Fig. 3).
Conservation of the marsupial Y chromosome
To investigate conservation of marsupial Y gene content, we searched for wallaby Y genes in Tasmanian devil, an Australian dasyurid marsupial that last shared an ancestor with macropods about 50 MYA (Nilsson et al. 2004). Sequence from both the X and Y copies of the 10 tammar X/Y shared genes were used in nucleotide BLAST queries of the male Tasmanian devil testis transcriptome (16,438 unique transcripts) (Murchison et al. 2010). BLAST hits were assigned as X or Y derived, depending on whether they had greater similarity to the Y query sequence, or the X query. Evidence of Tasmanian devil X- and Y-derived sequences was obtained for all 10 tammar X–Y shared genes. This suggests an ancestral core of at least 10 X–Y shared genes that have been conserved on the Y chromosome in Australian marsupials.
BLAST searches revealed no evidence of a Tasmanian devil SPATA2Y ortholog, suggesting that retrotransposition of the autosomal SPATA2 to the Y is a recent event in the macropod ancestor. This is consistent with the high-sequence identity (95%) between SPATA2 and SPATA2Y (Table 3).
Discussion
We have sequenced a total of ∼1 Mb of the euchromatic short arm (∼10 Mb) of the tammar wallaby Y, and discovered five novel genes (Table 1), bringing the number of known Y genes to 10 in this species. Nine of these genes (KDM5D was not detected on the BACs sequenced herein) were detected in ∼1 Mb of sequence, so if the remainder of the short arm was devoid of genes, the gene-rich region of the tammar wallaby Y could be very compact. All tammar Y genes have a partner on the tammar X with an ortholog on the human X.
Unlike the testis-specific genes SRY, RBMY, UBE1Y, and ATRY described previously on the marsupial Y, and most genes on the human Y, the newly discovered tammar Y genes are all ubiquitously expressed, although most at a much lower level than their X homologs (Fig. 3). We found evidence of Y-specific orthologs in the Tasmanian devil for all 10 genes, complementing previous descriptions of SRY, RBMY, and UBE1Y from another dasyurid, the stripe-face dunnart (Foster et al. 1992; Mitchell et al. 1992; Delbridge et al. 1997). This establishes that at least 10 genes (SRY, RBMY, UBE1Y, ATRY, KDM5D, PHF6Y, HUWE1Y, HCFC1Y, RPL10Y, MECP2Y) lay on the Y chromosome of the common ancestor of Australian marsupials ∼45 MYA. Because the marsupial Y chromosome has been evolving independently from the eutherian Y for ∼148 MY, these findings have important implications for our understanding of mammalian Y chromosome evolution.
Evolutionary history of therian mammal Y genes
Here, we estimate synonymous and nonsynonymous substitutions between nine tammar XY shared genes, and between human and tammar X orthologs. Ka/Ks ratios of <1 (Table 3) for all marsupial XY gene pairs indicate that they are under purifying selection (suggestive of a functional Y homolog, as observed for SRY in eutherian mammals; (King et al. 2007), and phylogenetic analyses indicate that some of these genes have had a complex evolutionary history.
For SOX3/SRY, RBMX/Y, and ATRX/Y the therian X orthologs form monophyletic clades (although with poor bootstrap support) (Fig. 2), and the Y copy/copies form a separate clade, suggesting that they were isolated from their X homologs before the marsupial/eutherian split. This is consistent with previous hypotheses (Foster et al. 1992; Delbridge et al. 1999; Pask et al. 2000; Waters et al. 2001): SRY is likely the testis determining factor in both marsupials and eutherians, a function that most certainly would have evolved only once. Likewise, RBMY probably acquired a testis-specific role before the therian radiation, and was retained on the Y in all taxa because of this role (possibly in spermatogenesis). This seems more plausible than independent selection for retention on the Y in multiple therian lineages.
In contrast to the SRY, RBMY, and ATRY topologies, the marsupial-specific HCFC1Y, MECP2Y, and HUWE1Y all group with their respective marsupial X homologs (Fig. 2), suggesting that they were isolated from their X homologs in the marsupial ancestor after the marsupial/eutherian split (Fig. 4). However, it cannot be excluded that gene conversion has occurred between these X and Y gametologues, as has been observed between X–Y gene pairs on the eutherian sex chromosomes (Pecon Slattery et al. 2000). Nevertheless, in all cases the wallaby and opossum X homologs are more closely related to each other than they are to the wallaby Y copy, so any gene conversion would have had to occur pre-marsupial radiation.
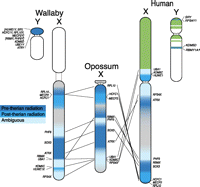
Two marsupial X chromosomes compared with the human X. Green is recently added to the eutherian sex chromosomes. No marsupial sequence was available for RPS4Y or KDM5D, so we could not predict when the X–Y gametologues diverged from each other. The three shades of blue represent X genes that diverged from their respective gametologues either pre-therian radiation, post-therian radiation, or unknown. Genes retained from the proto-Y on the tammar wallaby and human Y chromosomes are shown. These results highlight the poor resolution of human X chromosome Stratum 1.
Topology of the RPL10X/Y and PHF6X/Y trees (although there is low bootstrap support) (Fig. 2) indicates a more complex evolutionary history. The marsupial Y homologs group with the eutherian X homologs rather than the marsupial X homologs. It is possible that the RPL10 and PHF6 gametologues were isolated from each other before the therian radiation, and that there was Y to X gene conversion of these genes in the eutherian ancestor (before the Y copy was ultimately lost). This left the marsupial Y copies more similar to the eutherian X copies than either are to the marsupial X homologs. Finally, marsupial UBE1Y groups with marsupial UBA1, and eutherian UBE1Y groups with eutherian UBA1. Therefore, both the marsupial and eutherian UBE1Y genes must have been independently isolated from their X partners, and subsequently selected for and retained on the Y. Alternatively, UBE1Y may have been isolated from UBA1 before the eutherian/marsupial split, and then undergone gene conversion in both the marsupial and eutherian ancestors.
These findings have broader implications for mammalian sex chromosome evolution. SRY, RBMY, ATRY, and RPL10Y (and perhaps PHF6Y), were all isolated from their X homologs before therian radiation, suggesting that the ancestral therian Y had undergone significant specialization and degradation early in its evolution. A minimal PAR was retained in the therian ancestor (which included HCFC1X/Y, MECP2X/Y, and HUWE1X/Y) that was subsequently lost (along with all X–Y recombination) in the marsupial ancestor, but rejuvenated by an autosomal addition that extended the PAR in the eutherian ancestor. These results also highlight the poor resolution of human X chromosome Stratum 1, of which genes located in the ancestral therian PAR (HCFC1, MECP2, and HUWE1) cannot be members (Fig. 4).
Lineage-specific genes on therian Y chromosomes imply independent Y degradation
In marsupials, all 10 Y genes so far discovered lie within the YCR (the region of the Y with homology with XCR), and are therefore derived from the ancient proto X-Y. In humans, only four genes lie in this region and most derive from the autosomal addition (YAR) (Waters et al. 2001). SRY, RBMY, and KDM5D are located on the Y in all therian mammals (including marsupials) studied so far. Because these genes have been retained on the Y for at least 148 million years, it is likely that they acquired selectable male-specific functions (probably in sex and spermatogenesis) before marsupials and eutherians diverged.
The tammar and devil Y chromosomes share six genes (ATRY, and the newly discovered PHF6Y, HUWE1Y, HCFC1Y, RPL10Y, MECP2Y) that are absent from the Y chromosome of any eutherian species. The presence of these genes on the marsupial, but not the eutherian Y (Fig. 4), implies that they were lost from the Y early in eutherian evolution. In contrast, just one gene with an XCR homolog (CUL4BY) appears to be Y-borne in at least one eutherian lineage (Laurasiatheria) (Murphy et al. 2006), but absent from the marsupial Y.
The retention of many more YCR genes in marsupials than in eutherians could mean that the marsupial Y chromosome degraded more slowly than the eutherian Y, or that these genes acquired selectable male-specific functions in marsupials, but not eutherians. A slower overall degeneration of the marsupial Y seems unlikely in view of the terminally differentiated X and Y in all marsupials (see Fernández-Donoso et al. 2010). The alternative hypothesis is favored by the low Ka/Ks ratios for all X–Y gene pairs (Table 3), which suggests that they are under purifying selection, probably because they gained male-specific functions. Indeed, many of these novel Y genes are good candidates for roles in early male marsupial development, because mutations in their X-borne human or mouse homologs have phenotypes including gonadal abnormalities or male infertility (Supplemental Table 2). Perhaps YCR genes could be lost from the eutherian Y, because the addition of the YAR supplied some genes with redundant functions in spermatogenesis.
If these novel genes on the marsupial Y do have male-specific functions, it is somewhat surprising that they are all expressed widely in tammar wallaby (albeit at much lower levels than their X gametologues, even in testis) (Fig. 3). However, the possibility remains that they are highly expressed in the testis at a stage of male development that we could not sample, or in a specialized subset of cells that we could not detect with quantitative PCR.
CNS and testis expression of X homologs; the raw material of Y genes
All of the active genes detected to date on the tammar Y have copies on the X chromosome, from which they evolved. Of the 10 tammar Y genes, all but two have human/mouse X homologs that are implicated in male reproduction, and all are implicated in human X-linked mental retardation (XLMR) syndromes or central nervous system (CNS) diseases. The striking functional bias of these X genes toward sexual differentiation (Supplemental Table 2), suggests that genes that already functioned in male reproduction (at least partially) provided much of the raw material for Y genes recruited into male-specific functions.
The bias for so-called “brains and balls” genes (an enigmatic class of genes over-represented on the human X that have functions in the brain, as well as the testis; Graves et al. 2002) among X homologs of Y genes is therefore particularly pronounced in marsupials (Graves 2006). The original autosome pair that differentiated into the X and Y may have already possessed genes with selectable functions in the brain and testis; or, alternatively, this autosomal pair coded for large multifunctional proteins that could be readily exapted into selectable functions in these two tissues (Graves and Peichel 2010).
Conclusions
Our finding of 10 genes on the marsupial Y, all with X-borne homologs, implies that the marsupial Y chromosome has suffered less gene loss than the orthologous region of the eutherian Y. SRY, RBMY, and ATRY were isolated from their X homologs before the marsupial/eutherian split, with only SRY and RBMY retained on the Y in eutherian mammals so far studied. HCFC1Y, MECP2Y, and HUWE1Y were isolated from their X homologs in the marsupial ancestor, so were part of the PAR when marsupials and eutherians diverged. UBE1Y may have been independently isolated from the X in both the marsupial and eutherian ancestor, and then selected for and retained on the Y chromosome in both lineages. The involvement of all of the human X-borne homologs (of marsupial Y genes) in brain and/or testis function suggests that “brains and balls” genes on the proto XY were those that were preferentially retained, and subsequently further specialized on the Y.
Methods
Generating Y-specific sequences
An alignment of human RBMX and tammar RBMX and RBMY sequences was performed in order to identify Y-specific regions within RBMY. Primer pairs were then designed within exons 2 and 9 and spanning the intron between exons 6 and 7. The primer pairs for SRY and ATRY were designed from existing tammar sequences within the single exon of SRY and within exons 7, 19, and 34 of ATRY.
ME_VIA and MEKBa BAC library screening
Male tammar wallaby BAC libraries (ME_VIA) (Sankovic et al. 2005); (Me_KBa: Arizona Genomica Institute) were screened with microdissected Y chromosome probes (Grutzner et al. 2002; Sankovic et al. 2006). ME_VIA was screened separately with PCR-generated probes for RBMY, SRY, and ATRY. Three tammar-specific RBMY probes (see Supplemental Table 3 for all primer sequences) were amplified from male tammar genomic DNA with the following cycling conditions: 2 min at 94°C; followed by 25 cycles of 94°C, 30 sec/60°C, 1 min/72°C, 2 min; then 72°C, 10 min. The SRY probe was amplified with the following cycling conditions: 2 min at 94°C; followed by 25 cycles of 94°C, 30 sec/54°C, 1 min/72°C, 2 min; then 72°C, 10 min. Finally, three ATRY PCR products were generated with the following cycling conditions: 2 min at 94°C; followed by 25 cycles of 94°C, 30 sec/54°C, 1 min/72°C, 2 min; then 72°C, 10 min. All products were sequenced to confirm their identity.
The SRY probe and the three RBMY and three ATRY PCR products were each pooled and [32P]dATP labeled using the Megaprime DNA labeling system (Amersham) according to the manufacturer's instructions. The three different pools were individually hybridized to male tammar BAC library filters at 60°C overnight (Sankovic 2005). Library filters were exposed to X-ray film for 48 h. Identity of positive clones was confirmed by PCR with the primer sets used to amplify the probes.
MEB1 BAC library screening
Tammar male-specific sequence was identified in the region containing and immediately flanking SRY using sequence from ME_VIA-112D12 (AC162780). Male specificity of sequence was ensured by masking for repeat sequences using RepeatMasker (Smit et al. 1996–2010) and by BLAST sequences to the trace archives of the female tammar genome (http://www.ncbi.nlm.nih.gov/BLAST/mmtrace.shtml) (Renfree et al. 2011). Two tammar-specific SRY PCR probes were then generated and used to screen the MEB1 male tammar BAC library using a two-step 3D PCR screening system as described by Asakawa et al. (1997). The cycling conditions for both rounds of PCR screening were as follows; 10 min at 96°C; followed by 45 cycles of 96°C, 30 sec/60°C, 30 sec/72°C, 30 sec; then 72°C, 10 min. Identity of positive clones was confirmed by PCR using a single colony as the template DNA.
MEFX Fosmid library screening
The X-chromosome-specific fosmid library (MEFX–RIKEN ASI) was constructed from DNA derived from flow-sorted X chromosomes using the same methodology as for the human chromosome 21 fosmid library CMF21 (Hattori et al. 2000; Park et al. 2000). Transcript sequences from the female tammar assembly in Ensembl for HCFC1X (ENSMEUT00000008232) and RPL10X (ENSMEUT00000013200) were used as query sequence in a megablast search against end sequences from a tammar X chromosome-specific fosmid library (A Fujiyama, A Toyoda, Y Kuroki, S Tatsumoto, and Y Sakaki, unpubl.). Tammar-specific PCR probes were generated for HCFC1X and RPL10X using these gene predictions from Ensembl and used to confirm the presence of HCFC1X and RPL10X in the isolated clones by PCR using a single colony as the template DNA.
Fluorescent in situ hybridization
Positive clones were mapped to the tammar Y chromosome by fluorescence in situ hybridization (FISH) on male metaphase chromosomes using Digoxigenin (DIG) and Biotin-labeled probes that were detected with antidigoxigenin-rhodamine and avidin-fluorescein following the procedure described by Koina et al. (2005). Image capture used a Zeiss Axioplan2 epifluorescence microscope and a thermoelectronically cooled charge-coupled device camera (RT Monochrome Spot, Diagnostic Instruments) with image analysis conducted using IPLab imaging software (Scanalytics, Inc.).
Sequencing
Five Y-specific ME_VIA BAC clones (ME_VIA-53A23, ME_VIA-80O22, ME_VIA-112D12, ME_VIA-54J2, and ME_VIA-97A3) were shotgun sequenced by the U.S. Department of Energy Joint Genome Institute (JGI). All other BAC clones (MEB1-052D08, MEB1-321E03, ME_KBa-20F12, ME_KBa-87K16, and ME_KBa-343E06) were sequenced using Sanger sequencing technology by shotgun approach at the National Institute of Genetics (NIG). Each BAC had a minimum of 1390 clones sequenced from both ends. At an average read length of 750 bp, this represents a minimum 10-fold coverage for BACs containing a 200-kb insert.
KaKs calculations
ClustalX was used to align coding regions of the wallaby X and Y homologs with the human X homolog. All alignments were manually inspected and refined. Gaps and poorly aligned sequence was removed, being careful not to introduce frame shifts. Alignments were used to calculate nucleotide identity between each X–Y shared gene. Synonymous (Ks) and nonsynonymous (Ka) substitution rates were calculated for each pairwise alignment using a maximum likelihood model averaging in KaKs_Calculator (β) (Zhang et al. 2006).
Phylogenetic analysis
ClustalX was used to align coding regions of wallaby X and Y homologs with homologs representative from each of the four superordinal placental mammal clades, and (where available) Monodelphis, platypus, chicken, and/or green anole, and Xenopus (see Supplemental Table 4 for all accession nos. of aligned sequences). Taxa vary between phylogenetic analyses because sequence was not available for each gene in all species. All alignments were manually inspected and refined. Protein alignments were used to manually refine (with care taken not to introduce frame-shifts) poorly aligned nucleotide sequence in MacClade (v4.08a). Alignments were analyzed using maximum likelihood in PAUP* (Swofford 2003). For each data set, model parameters were estimated using jModeltest (Posada 2008), and heuristic searches were performed with 10 random taxon addition replicates and TBR branch swapping. Bootstrap values were estimated by running 500 replicates of the above analysis for each data set. For alignments and model parameters see nexus files in the Supplemental Information 1.
qPCR
Tammar tissues were collected under The Australian National University Animal Experimentation Ethics Committee proposal numbers R.CG.11.06 and R.CG.14.08. RNA was extracted from two male tammar wallaby tissue series (brain cortex, kidney, liver, lung, spleen, testis) with a GenElute Mammalian Total RNA Miniprep Kit (Sigma) according to the manufacturer's instructions. RNA was also extracted from ovarian tissue of a female and from male fibroblast cell lines. Reverse transcriptions were performed with SuperScript III First-Strand Synthesis System for RT–PCR (Invitrogen) according the manufacturer's instructions.
Suitable qPCR primers, producing a 100–150-bp product, were designed for each X/Y shared gene and the control gene. All primer pairs were tested on male and female genomic DNA, as well as testis cDNA in a PCR using the same cycling conditions as described for qPCR below. All primers generated single PCR products from male-derived DNA of the expected size, and their identity was confirmed by direct sequencing. No products were observed using Y-specific primers to amplify from female-derived cDNA or genomic DNA. The amplification efficiencies and the specificity of each primer pair were tested in a qPCR reaction from the first strand synthesis of testis RNA. The melting curve produced from each reaction was consistent with a single PCR product from each primer pair. qPCR reactions were set up in triplicate with the QuantiTect SYBR Green PCR system (QIAGEN), and reactions run on a Corbett Rotorgene 3000 (QIAGEN). Cycling conditions were as follows: 15 min at 95°C; followed by 45 cycles of 94°C, 15 sec/58°C, 20 sec/72°C, 20 sec; followed by a 55°C–99°C melt analysis to check product specificity. We tested expression of ATRX/Y, RBMX/Y, UBA1/UBE1Y, PHF6X/Y, and HUWE1X/Y on a tissue series from animal 1, and HCFC1X/Y, MECP2X/Y, and RPL10X/Y on tissues from animal 2. All expression levels were normalized to the autosomal gene GAPDH using comparative quantitation software supplied by Rotorgene.
Gene nomenclature
Due to alterations in nomenclature, clarification is required for the following genes.
Human nomenclature
-
KDM5C: previously known as SMCX, JARID1C, aliases DXS1272E, XE169
-
KDM5D: previously known as SMCY, JARID1D, HY, HYA, alias KIAA0234
-
UBA1: previously known as UBE1, A1S9T, GXP1, aliases UBE1X, POC20
Mouse nomenclature
-
Uba1: previously known as A1s9, Sbx, Ube-1, Ube1x
-
Ube1y: previously known as A1s9Y-1, Sby, Sby, Ube-2, Ube1y-1
Data access
The sequence data described in this study are available at GenBank under accession numbers AC162773, AC162779, AC162780, HM191417, AC162776, AC162777, JF810456, JF810457, JF810458, and JF810459.
Acknowledgments
We are especially thankful to the technical staff of the Comparative Genomics Laboratory NIG and the RIKEN Advanced Science Institute, the Victorian Institute of Animal Sciences (VIAS) for construction of the ME_VIA library, Ke-Jun Wei for curation of the ME_VIA tammar BAC library, and Anthony Papenfuss for bioinformatic tools used during this project. This project was funded by grants from the Australian Research Council (to J.A.M.G. and P.D.W.). Part of this work was performed under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, contract No. DE-AC02-05CH11231 with the University of California, Lawrence Berkeley National Laboratory. Part of this work was supported by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Area “Comparative Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to A.F. and Y.K.).
Authors' contributions: V.J.M. and N.S. performed the library screening, clone localizations, and characterization of the sequenced BACs. D.O. and P.D.W. conducted the phylogenetic analyses. J.L.B. coordinated the ME_VIA BAC sequencing, assembly, gene annotation, and interpretation. A.T. coordinated the MEB1 BAC sequencing and assembly. V.J.M., K.S.J., and P.D.W. performed the expression analysis. M.L.D. supervised the ME_VIA library work and contributed technical expertise to the expression analysis. A.F, Y.K., and A.T. carried out end sequencing of the MEFX library as part of the KanGO project. Y.K. and A.F. supervised the MEB1 library work and provided MEB1 and MEFX clone samples. V.J.M. and P.D.W. drafted and revised the manuscript. M.L.D., A.J.P., Y.K., J.L.B., D.O., and M.B.R. contributed to drafts of the manuscript. J.A.M.G. conceived the study and contributed to its design and coordination, and to the preparation and revision of the manuscript. P.D.W. contributed to the design and coordination of the study, and was responsible for revision of the manuscript after review.
Footnotes
-
↵11 Corresponding author.
E-mail Paul.waters{at}anu.edu.au.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.120790.111.
- Received January 12, 2011.
- Accepted November 16, 2011.
- Copyright © 2012 by Cold Spring Harbor Laboratory Press
Freely available online through the Genome Research Open Access option.