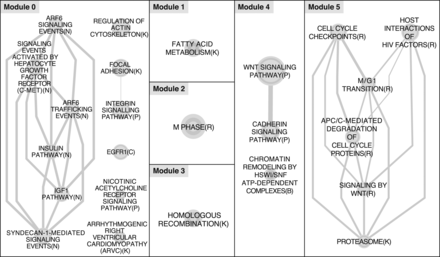
Pathway enrichment analysis of genes with monoallelic expression (MAE) established by loss-of-heterozygosity (LOH) events. Gene networks were inferred using Reactome Functional Interaction software (Wu et al. 2010) within the Cytoscape (Smoot et al. 2011) plug-in. LOH-induced MAE genes were used in the analysis and subsequently clustered into modules. At a false discovery rate (FDR) of 0.05, significantly enriched pathways included Modules 0–5. Shown are the Enrichment Map (Merico et al. 2010) networks generated for the significant pathways (Supplemental Table S13), highlighting the interactions between pathways identified within each of the six modules.











