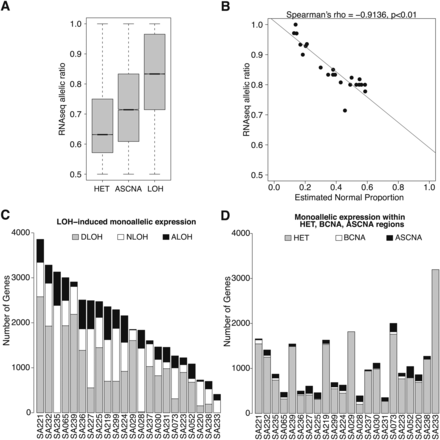
Integrative analysis of APOLLOH results and transcriptome RNA-seq expression. (A) The distribution of transcriptome RNA-seq symmetric allelic ratios that fall within HET, ASCNA, and LOH predicted regions are significantly different (pairwise Wilcoxon one-tailed test, p < 0.01). (B) The median symmetric allelic ratio of RNA-seq data within predicted LOH segments for each sample, represented as a point, strongly negatively correlated (Spearman's rho = −0.91) with estimated normal proportion parameter s (first principal component line is shown). (C,D) Distribution of the number of monoallelic expressed genes within genomic loss-of-heterozygosity (LOH), heterozygous (HET), and allele-specific copy number amplification (ASCNA) regions in 23 breast cancer samples. (C) The number of MAE genes established by LOH events are categorized into deletion (DLOH), copy neutral (NLOH), and amplification (ALOH) and sorted by total LOH in descending order. (D) The number of genes with MAE that overlapped genomic HET, balanced CNA (BCNA), and ASCNA regions are shown in same sorted order as in C.











