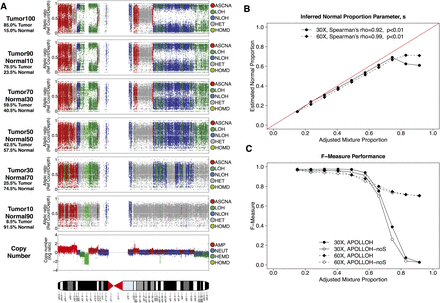
Tumor-normal sampling admixture experiment. Nine mixture proportions generated by sampling reads from the tumor and normal BAM files were analyzed (see Methods). (A) APOLLOH results are shown for chromosome 9 of mixture proportions of 0.09, 0.26, 0.43, 0.60, and 0.77 tumor reads sampled to 30×. (Tumour100) Results from the original tumor sample. (B) The normal proportion parameter s inferred by APOLLOH was significantly correlated (Spearman's rho = 0.92) with the mixture proportions of 0.1–1.0 (increments of 0.1) at 30× and 60×. (C) The F-Measure performance of APOLLOH and APOLLOH-noS (not accounting for normal contamination) for 30× and 60× admixtures was evaluated using Affymetrix SNP6.0 data as ground truth.











