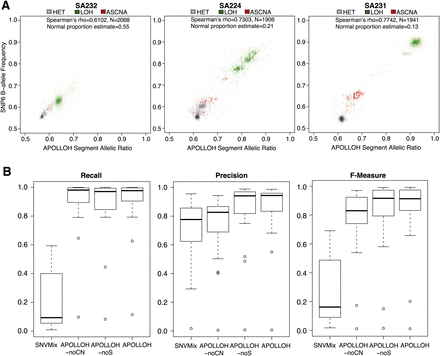
Comparison and evaluation of APOLLOH results using data from Affymetrix SNP6.0 genotyping arrays as the benchmark. (A) Initial benchmarking by comparing WGSS-derived allelic ratios and SNP6 B-allele frequencies. Three samples are shown with LOH clusters centered at locations reflecting APOLLOH normal contamination estimation. (B) For the 23 TNBC samples, precision, recall, and F-measure metrics were computed for LOH predictions from each APOLLOH model variant and SNVMix using OncoSNP (Yau et al. 2010) predictions (from SNP6 data) as the ground truth.











